
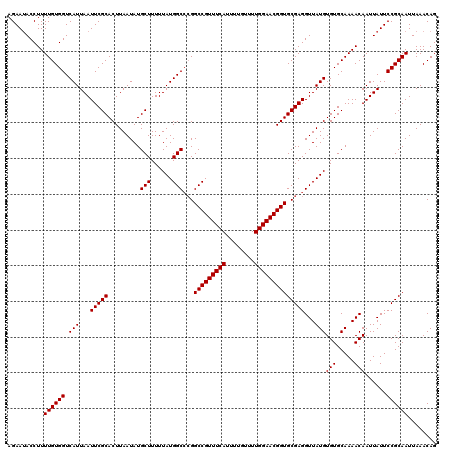
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,594,688 – 20,594,871 |
| Length | 183 |
| Max. P | 0.921265 |

| Location | 20,594,688 – 20,594,802 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.56 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -31.60 |
| Energy contribution | -32.40 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20594688 114 - 23771897 UUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAGUUUGCACUUCGCCAA------GAGAGAGCGGGCAGGUCCA .......((((((((((((((........)))))))))(((((((....(((.....)))........((((.........)))))))))))...------.......)))))....... ( -36.20) >DroSec_CAF1 39406 114 - 1 UUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAGUUUGCACUUCGCCAA------GAGAGAGCGGGCAGGUCCA .......((((((((((((((........)))))))))(((((((....(((.....)))........((((.........)))))))))))...------.......)))))....... ( -36.20) >DroAna_CAF1 61882 120 - 1 UUUUAUGGCCAGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAGUUUGCACUUCGCCAAGAAAAAGAGAGAGCGACCAGGUCCA ......((((.((.(((((..((((.((((((...(((((((.(((...((((.((........)).))))....)))...)))))))...))))))..))))..))))).)).)))).. ( -32.60) >DroSim_CAF1 32858 114 - 1 UUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAGUUUGCACUUCGCCAA------GAGAGAGCGGGCAGGUCCA .......((((((((((((((........)))))))))(((((((....(((.....)))........((((.........)))))))))))...------.......)))))....... ( -36.20) >DroEre_CAF1 32096 113 - 1 UUUUAUGGC-CGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAGUUUGCACUUCGCCAA------GAGAGAGCGGGCAGGUCCA ......(((-(.(((((((((........)))))))))(((..((....(((.....)))...))..))).........((((((.((((.....------)).)).)))))).)))).. ( -33.00) >consensus UUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAGUUUGCACUUCGCCAA______GAGAGAGCGGGCAGGUCCA .......((((((((((((((........)))))))))(((((((....(((.....)))........((((.........)))))))))))................)))))....... (-31.60 = -32.40 + 0.80)


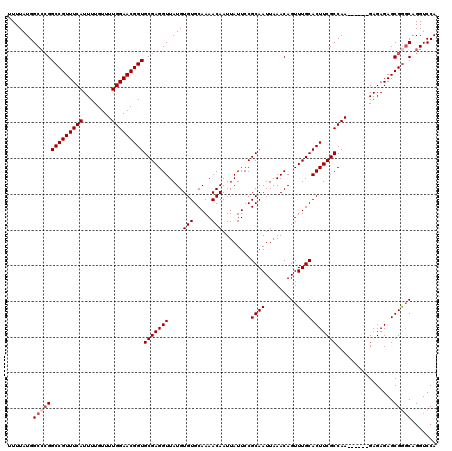
| Location | 20,594,722 – 20,594,842 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -29.24 |
| Energy contribution | -29.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20594722 120 - 23771897 AGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAG ..........((((((....(((((.((((.............((((((((((((((((((........))))))))).)).))))))).)))).....)))))..))))))........ ( -30.94) >DroSec_CAF1 39440 120 - 1 AGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAG ..........((((((....(((((.((((.............((((((((((((((((((........))))))))).)).))))))).)))).....)))))..))))))........ ( -30.94) >DroAna_CAF1 61922 120 - 1 AGAAAACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCAGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAG .............(((((((......(((.......))).....))))))).(((((((((........)))))))))(((..((....(((.....)))...))..))).......... ( -29.70) >DroSim_CAF1 32892 120 - 1 AGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAG ..........((((((....(((((.((((.............((((((((((((((((((........))))))))).)).))))))).)))).....)))))..))))))........ ( -30.94) >DroEre_CAF1 32130 119 - 1 AGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGC-CGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAG .(((((..(((((((..(((...(((((.........(((......)))-..(((((((((........))))))))))))))...)))..)))))))....)))))............. ( -29.40) >consensus AGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGCGAGGUUAUGUGUGCAAAACAAUUAUUCCGCAAUUAAACAG ..........((((((.(((...(((((.........(((......)))...(((((((((........))))))))))))))...)))(((.....)))......))))))........ (-29.24 = -29.24 + 0.00)

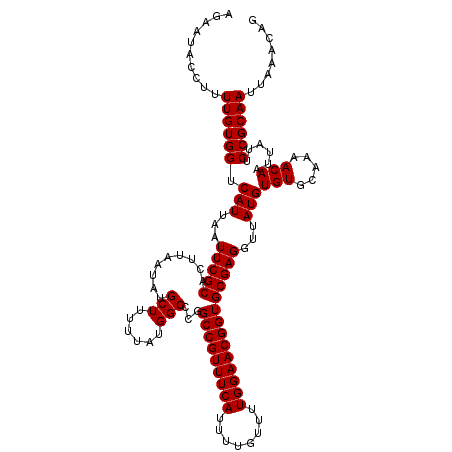
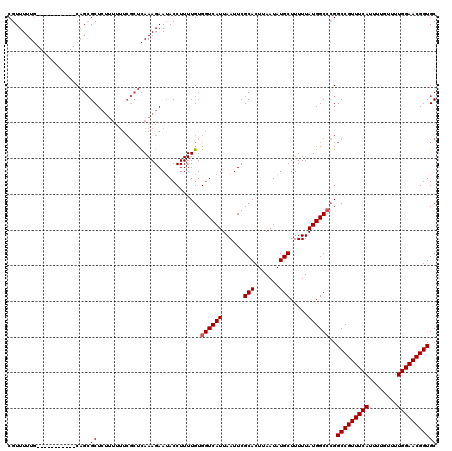
| Location | 20,594,762 – 20,594,871 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.12 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -19.54 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20594762 109 - 23771897 CGUUUUUG-----------CAGCGCUCUUUUUUCGCUCAAAGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGC ..((((((-----------.((((.........))))))))))........((.((((((......(((.......))).....))))))))(((((((((........))))))))).. ( -27.70) >DroSec_CAF1 39480 109 - 1 CGUUUUUG-----------CAGCGCUCUUUUUUCGCUCAAAGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGC ..((((((-----------.((((.........))))))))))........((.((((((......(((.......))).....))))))))(((((((((........))))))))).. ( -27.70) >DroAna_CAF1 61962 110 - 1 CGUUUUUCUUCCUCAACGGCUGAGCU----------UUAAAGAAAACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCAGGCCGUUUCAUUUUGUUUUGGAACGGUGC .(.(((((((.((((.....))))..----------...))))))).).....(((((((......(((.......))).....))))))).(((((((((........))))))))).. ( -27.30) >DroSim_CAF1 32932 109 - 1 CGUUUUUG-----------CAGCGCUCUUUUUUCGCUCAAAGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGC ..((((((-----------.((((.........))))))))))........((.((((((......(((.......))).....))))))))(((((((((........))))))))).. ( -27.70) >DroEre_CAF1 32170 108 - 1 CGUUUUUG-----------CAGCGCUCUUUUUUCGCUCAAAGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGC-CGGCCGUUUCAUUUUGUUUUGGAACGGUGC ..((((((-----------.((((.........))))))))))..........(((((((......(((.......))).....)))))-))(((((((((........))))))))).. ( -28.20) >consensus CGUUUUUG___________CAGCGCUCUUUUUUCGCUCAAAGAAUACCUUUUGUGGUCAUUAAUUCGCACUUAAUAUGCUUUUUAUGGCCCGGCCGUUUCAUUUUGUUUUGGAACGGUGC .......................((.............................((((((......(((.......))).....))))))..(((((((((........))))))))))) (-19.54 = -19.74 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:08 2006