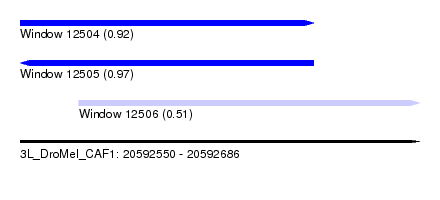
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,592,550 – 20,592,686 |
| Length | 136 |
| Max. P | 0.970864 |

| Location | 20,592,550 – 20,592,650 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -24.04 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20592550 100 + 23771897 GC--------------------UGCACUCAAUAUUCAAUUUUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGAC ..--------------------(((((...............((((((......))))))..(((..(((.......)))))))))))......((((..........))))........ ( -21.80) >DroSec_CAF1 37229 100 + 1 GC--------------------UGCACUCAAUAUUCAAUUUUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGAC ..--------------------(((((...............((((((......))))))..(((..(((.......)))))))))))......((((..........))))........ ( -21.80) >DroAna_CAF1 60301 117 + 1 CCGCUUACCGCAGCUUACUGCUUGCACUCAAUAUUCAAUUUUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGUUGGCGUGCAAAAAUAAGCCAAAGCCUUGGGGCCGGAGA--- (((((((..((((....))))((((((...............((((((......))))))..(((..((........)).)))))))))...)))))....(((....))).))...--- ( -31.60) >DroSim_CAF1 30724 100 + 1 GC--------------------UGCACUCAAUAUUCAAUUUUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGAC ..--------------------(((((...............((((((......))))))..(((..(((.......)))))))))))......((((..........))))........ ( -21.80) >DroEre_CAF1 30393 100 + 1 GC--------------------UGCACUCAAUAUUCAAUUUUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAGCCCAAAAGGCUCAAGAGAC ..--------------------....(((.............((((((......))))))(.((((((((.......)))(((..((........))...)))...))))).)..))).. ( -23.20) >consensus GC____________________UGCACUCAAUAUUCAAUUUUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGAC ......................(((((...............((((((......))))))..(((..(((.......))))))))))).......(((..........)))......... (-19.74 = -19.94 + 0.20)

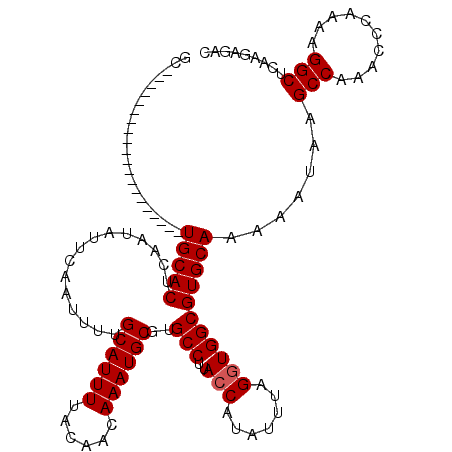
| Location | 20,592,550 – 20,592,650 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.41 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -24.86 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20592550 100 - 23771897 GUCUCUUGAGCCUUUUGGGUUUGGCUUAUUUUUGCACGCCACCUAAAUAUGGUAAGGCACGCAUUUGUUGUAAAAUGCAAAAUUGAAUAUUGAGUGCA--------------------GC ......((((((..........))))))...((((((((((((.......)))..)))..((((((......))))))...............)))))--------------------). ( -28.20) >DroSec_CAF1 37229 100 - 1 GUCUCUUGAGCCUUUUGGGUUUGGCUUAUUUUUGCACGCCACCUAAAUAUGGUAAGGCACGCAUUUGUUGUAAAAUGCAAAAUUGAAUAUUGAGUGCA--------------------GC ......((((((..........))))))...((((((((((((.......)))..)))..((((((......))))))...............)))))--------------------). ( -28.20) >DroAna_CAF1 60301 117 - 1 ---UCUCCGGCCCCAAGGCUUUGGCUUAUUUUUGCACGCCAACUAAAUAUGGUAAGGCACGCAUUUGUUGUAAAAUGCAAAAUUGAAUAUUGAGUGCAAGCAGUAAGCUGCGGUAAGCGG ---...((((((....))))...(((((((.(((((((((.((((....))))..)))..((((((......))))))...............))))))((((....))))))))))))) ( -38.00) >DroSim_CAF1 30724 100 - 1 GUCUCUUGAGCCUUUUGGGUUUGGCUUAUUUUUGCACGCCACCUAAAUAUGGUAAGGCACGCAUUUGUUGUAAAAUGCAAAAUUGAAUAUUGAGUGCA--------------------GC ......((((((..........))))))...((((((((((((.......)))..)))..((((((......))))))...............)))))--------------------). ( -28.20) >DroEre_CAF1 30393 100 - 1 GUCUCUUGAGCCUUUUGGGCUUGGCUUAUUUUUGCACGCCACCUAAAUAUGGUAAGGCACGCAUUUGUUGUAAAAUGCAAAAUUGAAUAUUGAGUGCA--------------------GC .....(..((((.....))))..).......((((((((((((.......)))..)))..((((((......))))))...............)))))--------------------). ( -29.30) >consensus GUCUCUUGAGCCUUUUGGGUUUGGCUUAUUUUUGCACGCCACCUAAAUAUGGUAAGGCACGCAUUUGUUGUAAAAUGCAAAAUUGAAUAUUGAGUGCA____________________GC ......((((((..........))))))....(((((((((((.......)))..)))..((((((......))))))...............)))))...................... (-24.86 = -25.30 + 0.44)

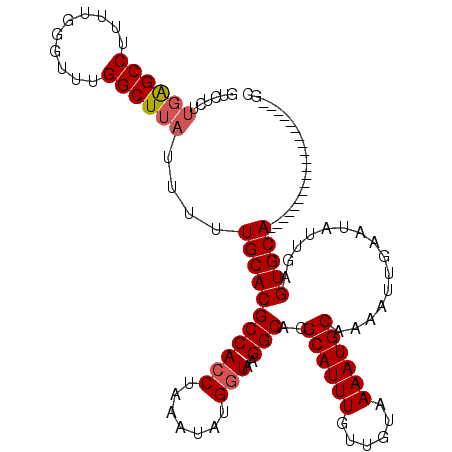

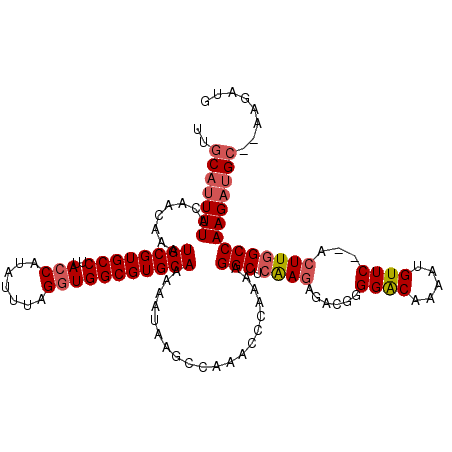
| Location | 20,592,570 – 20,592,686 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -22.28 |
| Energy contribution | -23.16 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20592570 116 + 23771897 UUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGACGGGGACAAAAUGUUC--ACUUGGCCAAGAUGC--AAGAUG (((((((((........((((((((..(((.......)))))))))))....................(((.((((......((((.....))))--.))))))))))))))--)).... ( -31.50) >DroSec_CAF1 37249 116 + 1 UUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGACGGGGACAAAAUGUUC--ACUUGGCCAAGAUGC--AAGAUG (((((((((........((((((((..(((.......)))))))))))....................(((.((((......((((.....))))--.))))))))))))))--)).... ( -31.50) >DroAna_CAF1 60341 115 + 1 UUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGUUGGCGUGCAAAAAUAAGCCAAAGCCUUGGGGCCGGAGA-----GGGCAAAAUGUUCCAAGUUGGCCAAGUUGGUUUGGGGA ..((((((......)))))).((((((.((........(((((............))))).(((....))).))..)-----))))).....(((((((..(((...)))..))))))). ( -32.60) >DroSim_CAF1 30744 116 + 1 UUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGACGGGGACAAAAUGUUC--ACUUGGCCAAGAUGC--AAGAUG (((((((((........((((((((..(((.......)))))))))))....................(((.((((......((((.....))))--.))))))))))))))--)).... ( -31.50) >DroEre_CAF1 30413 116 + 1 UUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAGCCCAAAAGGCUCAAGAGACGGGGACAAAAUGUUC--ACUUGGCCAAGAAGC--AAGAUG ((((.((((........((((((((..(((.......))))))))))).......((((((((.....)))...........((((.....))))--..))))).)))).))--)).... ( -29.30) >consensus UUGCAUUUUACAACAAAUGCGUGCCUUACCAUAUUUAGGUGGCGUGCAAAAAUAAGCCAAACCCAAAAGGCUCAAGAGACGGGGACAAAAUGUUC__ACUUGGCCAAGAUGC__AAGAUG ..(((((((........((((((((..(((.......)))))))))))....................(((.((((......((((.....))))...))))))))))))))........ (-22.28 = -23.16 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:03 2006