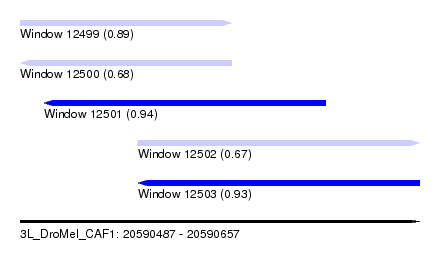
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,590,487 – 20,590,657 |
| Length | 170 |
| Max. P | 0.936094 |

| Location | 20,590,487 – 20,590,577 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -25.91 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20590487 90 + 23771897 AUCAUGCUGAAUGGAGGGGUGAUGUCAGGUAGUCCCGGUACGAGUUUUGGGCUCUAGAGGAUGGGCCAGGAUGGCAACUAAGUUGCCAUA (((((.((.......)).)))))....(((.(((((.....((((.....))))..).))))..)))...((((((((...)))))))). ( -27.00) >DroSec_CAF1 35225 90 + 1 AUCAUGCUGAAUGGAGGGGCGAUGUCAGGUAGCCCCGGUUCGGGUUUUGGGCUCUAGAGGAUGGGCCAGGAUGGCAACUAAGUUGCCAUA ......((((((...(((((...........))))).))))))......(((((........)))))...((((((((...)))))))). ( -34.70) >DroSim_CAF1 28728 90 + 1 AUCAUGCUGAAUGGAGGGGCGAUGUCAGGUAGCCCCGGUUCGGGUUUUGGGCUCUAGAGGAUGGGCCAGGAUGGCAACUUAGUUGCCAUA ......((((((...(((((...........))))).))))))......(((((........)))))...((((((((...)))))))). ( -34.70) >DroEre_CAF1 28390 86 + 1 AUGAAGCUGAAUGGAGG-GCGAUGCCAGGUAGCCUCUGUUCGAGAUUCGGGCUCUAGAG---AGGCCAGGAUGGCAACUAAGUUGCCAUG ..(((.(((((((((((-((........))..))))))))).)).)))((.(((....)---)).))...((((((((...)))))))). ( -31.80) >consensus AUCAUGCUGAAUGGAGGGGCGAUGUCAGGUAGCCCCGGUUCGAGUUUUGGGCUCUAGAGGAUGGGCCAGGAUGGCAACUAAGUUGCCAUA ......((((((...(((((...........))))).))))))......(((((........)))))...((((((((...)))))))). (-25.91 = -26.60 + 0.69)



| Location | 20,590,487 – 20,590,577 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 89.81 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -16.40 |
| Energy contribution | -15.59 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20590487 90 - 23771897 UAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACUCGUACCGGGACUACCUGACAUCACCCCUCCAUUCAGCAUGAU ..(((((((...)))))))((((((..(........(((.......))))..))))))....((((..............))))...... ( -17.14) >DroSec_CAF1 35225 90 - 1 UAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACCCGAACCGGGGCUACCUGACAUCGCCCCUCCAUUCAGCAUGAU .((((((((...))))))))...(((.(........).)))........(((..(((((...........)))))....)))........ ( -22.00) >DroSim_CAF1 28728 90 - 1 UAUGGCAACUAAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACCCGAACCGGGGCUACCUGACAUCGCCCCUCCAUUCAGCAUGAU .((((((((...))))))))...(((.(........).)))........(((..(((((...........)))))....)))........ ( -22.00) >DroEre_CAF1 28390 86 - 1 CAUGGCAACUUAGUUGCCAUCCUGGCCU---CUCUAGAGCCCGAAUCUCGAACAGAGGCUACCUGGCAUCGC-CCUCCAUUCAGCUUCAU .((((((((...))))))))..((((((---((...(((.......)))....))))))))...(((...))-)................ ( -24.40) >consensus UAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACCCGAACCGGGGCUACCUGACAUCGCCCCUCCAUUCAGCAUGAU .((((((((...))))))))...(((.(........).))).....(((.....))).....((((..............))))...... (-16.40 = -15.59 + -0.81)

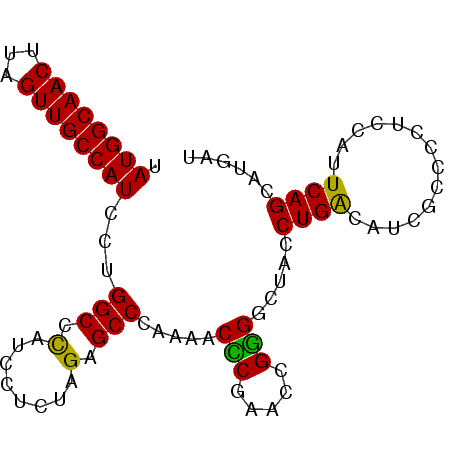

| Location | 20,590,497 – 20,590,617 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -26.18 |
| Energy contribution | -27.66 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20590497 120 - 23771897 UUCAGGCCUCCAUAACCUUGCGAAGGGGUUUAGGUUAGGGUAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACUCGUACCGGGACUACCUGACAUCACCCCUCCAU .(((((..(((.......(((((..(((((((((...((((((((((((...))))))))....))))..)))...)))))).....)))))...)))...))))).............. ( -38.20) >DroSec_CAF1 35235 120 - 1 UUCAGGCCUCCAUAACCUUGCGAAUGGGUUUAGGUUAGGGUAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACCCGAACCGGGGCUACCUGACAUCGCCCCUCCAU (((.((..((((......)).)).((((((((((...((((((((((((...))))))))....))))..)))...)))))))...)).)))..(((((...........)))))..... ( -39.40) >DroAna_CAF1 58281 104 - 1 UCCAGGCCUUUAUAACCUUGCGAAUG-GCUUAGGGAAGGGUAUGGCAACUUUGUUGCCAAUCU--------UUCUUGAUCC---AACGUGAAUA-GGCCUACCUGCCAUUGCUCC---AU ...((((((........(..((..((-(.((((((((((...(((((((...))))))).)))--------))))))).))---).))..)..)-)))))...............---.. ( -33.50) >DroSim_CAF1 28738 120 - 1 UUCAGGCCUCCAUAACCUUGCGAAUGGGUUUAGGUUAGGGUAUGGCAACUAAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACCCGAACCGGGGCUACCUGACAUCGCCCCUCCAU (((.((..((((......)).)).((((((((((...((((((((((((...))))))))....))))..)))...)))))))...)).)))..(((((...........)))))..... ( -39.40) >DroEre_CAF1 28400 115 - 1 UUCAGGCCUCCAUAACCUUGCGAAUG-GUUUGGGUUAGGGCAUGGCAACUUAGUUGCCAUCCUGGCCU---CUCUAGAGCCCGAAUCUCGAACAGAGGCUACCUGGCAUCGC-CCUCCAU ....(((..(((.((((........)-))))))(((((((.((((((((...)))))))).)((((((---((...(((.......)))....))))))))))))))...))-)...... ( -42.00) >consensus UUCAGGCCUCCAUAACCUUGCGAAUGGGUUUAGGUUAGGGUAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGCCCAAAACCCGAACCGGGGCUACCUGACAUCGCCCCUCCAU .(((((((((..((((((.((......))..))))))((((.(((((((...)))))))..((((........)))).))))............))))...))))).............. (-26.18 = -27.66 + 1.48)


| Location | 20,590,537 – 20,590,657 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -24.08 |
| Energy contribution | -26.48 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20590537 120 + 23771897 GCUCUAGAGGAUGGGCCAGGAUGGCAACUAAGUUGCCAUACCCUAACCUAAACCCCUUCGCAAGGUUAUGGAGGCCUGAAAAAAGAAAUUCUGAAUAAUUCGUGAUCUGCAGGCGCAAAA .(((((((((.((((..(((((((((((...))))))))..)))..))))....))))(....)....)))))(((((.....(((...(((((.....))).))))).)))))...... ( -34.30) >DroSec_CAF1 35275 120 + 1 GCUCUAGAGGAUGGGCCAGGAUGGCAACUAAGUUGCCAUACCCUAACCUAAACCCAUUCGCAAGGUUAUGGAGGCCUGAAAAAAGAAAUUCUGAAUAAUUCGUGAUCUGCAGGCGCAAAA ((.((...(((((((..(((((((((((...)))))))).......)))...)))))))(((.(((((((((.....(((........))).......)))))))))))).)).)).... ( -36.31) >DroAna_CAF1 58314 111 + 1 GAUCAAGAA--------AGAUUGGCAACAAAGUUGCCAUACCCUUCCCUAAGC-CAUUCGCAAGGUUAUAAAGGCCUGGAAAAAGAAAUUCUGAAUAAUUCGUGAUCCGCAGGCGCAAAA (((((.(((--------....(((((((...))))))).....((((....((-.....)).(((((.....))))))))).................))).))))).((....)).... ( -24.50) >DroSim_CAF1 28778 120 + 1 GCUCUAGAGGAUGGGCCAGGAUGGCAACUUAGUUGCCAUACCCUAACCUAAACCCAUUCGCAAGGUUAUGGAGGCCUGAAAAAAGAAAUUCUGAAUAAUUCGUGAUCUGCAGGCGCAAAA ((.((...(((((((..(((((((((((...)))))))).......)))...)))))))(((.(((((((((.....(((........))).......)))))))))))).)).)).... ( -36.31) >DroEre_CAF1 28439 116 + 1 GCUCUAGAG---AGGCCAGGAUGGCAACUAAGUUGCCAUGCCCUAACCCAAAC-CAUUCGCAAGGUUAUGGAGGCCUGAAAAAAGAAAUUCUGAAUAAUUCGUGAUCUGCAGGCGCAAAA (((.((((.---(((((.((((((((((...)))))))).)).....((((((-(........)))).))).)))))............(((((.....))).))))))..)))...... ( -33.00) >consensus GCUCUAGAGGAUGGGCCAGGAUGGCAACUAAGUUGCCAUACCCUAACCUAAACCCAUUCGCAAGGUUAUGGAGGCCUGAAAAAAGAAAUUCUGAAUAAUUCGUGAUCUGCAGGCGCAAAA .(((((.....((((..(((((((((((...))))))))..)))..)))).......((....))...)))))(((((.....(((...(((((.....))).))))).)))))...... (-24.08 = -26.48 + 2.40)


| Location | 20,590,537 – 20,590,657 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -28.16 |
| Energy contribution | -28.00 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20590537 120 - 23771897 UUUUGCGCCUGCAGAUCACGAAUUAUUCAGAAUUUCUUUUUUCAGGCCUCCAUAACCUUGCGAAGGGGUUUAGGUUAGGGUAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGC ......(((((.(((....(((((......)))))....))))))))............((..(((((....(((((((..((((((((...)))))))))))))))...)))))...)) ( -36.20) >DroSec_CAF1 35275 120 - 1 UUUUGCGCCUGCAGAUCACGAAUUAUUCAGAAUUUCUUUUUUCAGGCCUCCAUAACCUUGCGAAUGGGUUUAGGUUAGGGUAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGC ......(((((.(((....(((((......)))))....))))))))(((...(((((.......)))))..(((((((..((((((((...))))))))))))))).........))). ( -33.50) >DroAna_CAF1 58314 111 - 1 UUUUGCGCCUGCGGAUCACGAAUUAUUCAGAAUUUCUUUUUCCAGGCCUUUAUAACCUUGCGAAUG-GCUUAGGGAAGGGUAUGGCAACUUUGUUGCCAAUCU--------UUCUUGAUC ......(((((.(((....(((((......)))))....))))))))........(((.((.....-))..)))(((((((.(((((((...)))))))))))--------)))...... ( -30.80) >DroSim_CAF1 28778 120 - 1 UUUUGCGCCUGCAGAUCACGAAUUAUUCAGAAUUUCUUUUUUCAGGCCUCCAUAACCUUGCGAAUGGGUUUAGGUUAGGGUAUGGCAACUAAGUUGCCAUCCUGGCCCAUCCUCUAGAGC ......(((((.(((....(((((......)))))....))))))))(((...(((((.......)))))..(((((((..((((((((...))))))))))))))).........))). ( -33.50) >DroEre_CAF1 28439 116 - 1 UUUUGCGCCUGCAGAUCACGAAUUAUUCAGAAUUUCUUUUUUCAGGCCUCCAUAACCUUGCGAAUG-GUUUGGGUUAGGGCAUGGCAACUUAGUUGCCAUCCUGGCCU---CUCUAGAGC ......(((((.(((...((((((((((.(((........)))(((.........)))...)))))-)))))(((((((..((((((((...))))))))))))))))---)).))).)) ( -35.50) >consensus UUUUGCGCCUGCAGAUCACGAAUUAUUCAGAAUUUCUUUUUUCAGGCCUCCAUAACCUUGCGAAUGGGUUUAGGUUAGGGUAUGGCAACUUAGUUGCCAUCCUGGCCCAUCCUCUAGAGC ......(((((.(((....(((((......)))))....))))))))(((..((((((.((......))..))))))(((.((((((((...))))))))))).............))). (-28.16 = -28.00 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:54:00 2006