
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,588,553 – 20,588,697 |
| Length | 144 |
| Max. P | 0.995276 |

| Location | 20,588,553 – 20,588,673 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -35.39 |
| Consensus MFE | -30.89 |
| Energy contribution | -30.89 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20588553 120 + 23771897 GGCGCAAUGUGCCCCCACAUGCAAAUUGGCUGCUCGACAAUCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACG (((((...)))))......(((((...((.(((..........))).)))))))((((((((((.......((((((..................))))))....))))))))))..... ( -36.67) >DroSec_CAF1 33299 120 + 1 GGCGCAAUGUGCCCCCACAUGCAAAUUGGCUGCUCGACAAUCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACG (((((...)))))......(((((...((.(((..........))).)))))))((((((((((.......((((((..................))))))....))))))))))..... ( -36.67) >DroAna_CAF1 54679 113 + 1 AUUCGAAUGUGCCCCCACAUGCAAAUUGGUUGCUCG-------GCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACG .(((..(((((....)))))((((...((.(((...-------))).)))))).((((((((((.......((((((..................))))))....))))))))))))).. ( -33.17) >DroSim_CAF1 26801 120 + 1 UGCGCAAUGUGCCCCCACAUGCAAAUUGGCUGCUCGACAAUCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACG ((((((.((((....))))))).....((.(((..........))).))..)))((((((((((.......((((((..................))))))....))))))))))..... ( -35.97) >DroEre_CAF1 26405 109 + 1 G-----------CCCCAGAUGCAAAUUGGCUGCUCGGCAAUCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACG (-----------(....(((((..((((.(.....).))))..)))))...)).((((((((((.......((((((..................))))))....))))))))))..... ( -34.47) >consensus GGCGCAAUGUGCCCCCACAUGCAAAUUGGCUGCUCGACAAUCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACG ...................(((((...((.(((..........))).)))))))((((((((((.......((((((..................))))))....))))))))))..... (-30.89 = -30.89 + 0.00)


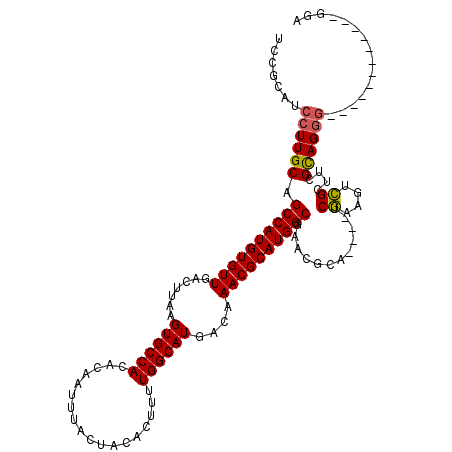
| Location | 20,588,593 – 20,588,697 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.35 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20588593 104 + 23771897 UCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACGCA----UCGAAGUCGCUUCGCAGGG------------GGA ......(((((((.((((((((((.......((((((..................))))))....))))))))))(((.((.----........)))))))))))------------).. ( -36.07) >DroSec_CAF1 33339 104 + 1 UCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACGCA----UCGAAGUCGCUUCGCAGGG------------GGA ......(((((((.((((((((((.......((((((..................))))))....))))))))))(((.((.----........)))))))))))------------).. ( -36.07) >DroAna_CAF1 54715 117 + 1 ---GCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACGCAUGCAUCCAAGUGGCCCAGUAGCUUCGGCUGCCCGUGUU ---(((....))).((((((((((.......((((((..................))))))....)))))))))).(((((......((....))....(((((....)))))..))))) ( -39.17) >DroSim_CAF1 26841 104 + 1 UCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACGCA----UCGAAGUCGCUUCGCAGGG------------GGA ......(((((((.((((((((((.......((((((..................))))))....))))))))))(((.((.----........)))))))))))------------).. ( -36.07) >DroEre_CAF1 26434 103 + 1 UCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACGCA----UCGAAGCCGCU-CGUAGGG------------AGA ......(((((((.((((((((((.......((((((..................))))))....))))))))))((.(((.----.....).)).)-)))))))------------).. ( -34.27) >consensus UCCGCAUCCUUGCAGCCAUGUGUUGACUUAAGUGCCACACAAUUUACUACACUUUUGGCAUGACAAACGCAUGGCGAACGCA____UCGAAGUCGCUUCGCAGGG____________GGA .......((((((.((((((((((.......((((((..................))))))....))))))))))............((....))....))))))............... (-29.51 = -29.35 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:55 2006