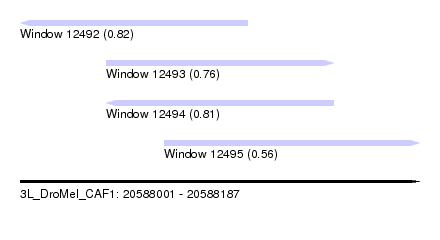
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,588,001 – 20,588,187 |
| Length | 186 |
| Max. P | 0.824098 |

| Location | 20,588,001 – 20,588,107 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.76 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20588001 106 - 23771897 AUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU-UCUGCCGU-CC------CACUCGCAC------CGAGUGGAUCAGUCUCGCUCCAUCUGCACACACACGGUUAUCAGGAACUUU ............((((..(((((......)))))..-...(((((-.(------((((((...------))))))).........((.......))......)))))....))))..... ( -27.10) >DroSec_CAF1 32784 102 - 1 AUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU-UCUGCCGU-CC------CACUCGCAC------CGAGUGGAUCUGUCUCGCUCCAUCUGCA----CACGGUUAUCAGGAACUUU ............((((..(((((......)))))..-...(((((-.(------((((((...------))))))).........((.......)).----.)))))....))))..... ( -27.10) >DroAna_CAF1 54297 103 - 1 AUUAACGUCACAUCCUU----GCUGU---UCGCCCUGUCUUCCACACCCAUACUCCUUCGCGG------CGAGGGGAUCUGGCUCUUUCCAUCUGCA----UUUGGUUAUCAGGAACUUU ......((((.(((((.----....(---(((((.((.....................)).))------))))))))).))))((((.(((......----..))).....))))..... ( -18.80) >DroSim_CAF1 26285 102 - 1 AUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU-UCUGCCGU-CC------CACUCGCAC------CGAGUGGAGCUGUCUCGCUCCAUCUGCA----CACGGUUAUCAGGAACUUU ............((((..(((((......)))))..-...(((((-..------.(((((...------)))))(((((......))))).......----.)))))....))))..... ( -28.20) >DroEre_CAF1 25917 108 - 1 AUUAACUUCACAUCCUCCGGCACAAUUAAGCGCCCU-UCUGCCGU-CC------CACUCGCACAUCCGACGGGUGCAUCUGCCCCGCUCCAUCUGCA----CGUGGUUAUCAGAAACUUU ..................(((.(......).))).(-((((....-.(------(((..(((.....((((((.((....)))))).))....))).----.))))....)))))..... ( -26.00) >consensus AUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU_UCUGCCGU_CC______CACUCGCAC______CGAGUGGAUCUGUCUCGCUCCAUCUGCA____CACGGUUAUCAGGAACUUU ............((((..(((((......)))))......((((..........((((((.........))))))..........((.......)).......))))....))))..... (-15.72 = -15.76 + 0.04)


| Location | 20,588,041 – 20,588,147 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -39.56 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.82 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20588041 106 + 23771897 UGAUCCACUCG------GUGCGAGUG------GG-ACGGCAGA-AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGC ..((((.(.((------((((((.((------((-...((...-...)).)))).))))))))).))))((((.((((.(((((((((..((....))..)))))))))))))...)))) ( -42.80) >DroSec_CAF1 32820 106 + 1 AGAUCCACUCG------GUGCGAGUG------GG-ACGGCAGA-AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGC ..((((.(.((------((((((.((------((-...((...-...)).)))).))))))))).))))((((.((((.(((((((((..((....))..)))))))))))))...)))) ( -42.80) >DroAna_CAF1 54333 107 + 1 AGAUCCCCUCG------CCGCGAAGGAGUAUGGGUGUGGAAGACAGGGCGA---ACAGC----AAGGAUGUGACGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUAGCAUAAUGAGCUCA ......((.((------((((......))...)))).))......((((..---(((..----.....)))..(((((.(((..((((..((....))..))))..)))))))).)))). ( -29.80) >DroSim_CAF1 26321 106 + 1 AGCUCCACUCG------GUGCGAGUG------GG-ACGGCAGA-AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGC ...(((.(.((------((((((.((------((-...((...-...)).)))).))))))))).))).((((.((((.(((((((((..((....))..)))))))))))))...)))) ( -41.30) >DroEre_CAF1 25953 112 + 1 AGAUGCACCCGUCGGAUGUGCGAGUG------GG-ACGGCAGA-AGGGCGCUUAAUUGUGCCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUACC ..((((..(((...((.((((((((.------((-.(((((..-..(((((......)))))....(((.....)))..))))))).))))))))))....)))..)))).......... ( -41.10) >consensus AGAUCCACUCG______GUGCGAGUG______GG_ACGGCAGA_AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGC .....((((((.........))))))..........((((......((.((......)).))................((((((((((..((....))..)))))))))).....)))). (-23.82 = -24.82 + 1.00)



| Location | 20,588,041 – 20,588,147 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -22.60 |
| Energy contribution | -22.44 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20588041 106 - 23771897 GCGACUCAUUAUGCCACCGAAAUGAAUGCAAAUCGGCGGCAUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU-UCUGCCGU-CC------CACUCGCAC------CGAGUGGAUCA ..(((.....(((((.((((..((....))..)))).)))))................(((((......)))))..-......))-)(------((((((...------))))))).... ( -32.20) >DroSec_CAF1 32820 106 - 1 GCGACUCAUUAUGCCACCGAAAUGAAUGCAAAUCGGCGGCAUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU-UCUGCCGU-CC------CACUCGCAC------CGAGUGGAUCU ..(((.....(((((.((((..((....))..)))).)))))................(((((......)))))..-......))-)(------((((((...------))))))).... ( -32.20) >DroAna_CAF1 54333 107 - 1 UGAGCUCAUUAUGCUACCGAAAUGAAUGCAAAUCGGCGGCAUUAACGUCACAUCCUU----GCUGU---UCGCCCUGUCUUCCACACCCAUACUCCUUCGCGG------CGAGGGGAUCU ..((.((...((((..((((..((....))..))))..))))..........(((((----(((((---..............................))))------)))))))).)) ( -23.51) >DroSim_CAF1 26321 106 - 1 GCGACUCAUUAUGCCACCGAAAUGAAUGCAAAUCGGCGGCAUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU-UCUGCCGU-CC------CACUCGCAC------CGAGUGGAGCU (((((.....(((((.((((..((....))..)))).)))))................(((((......)))))..-......))-)(------((((((...------))))))).)). ( -34.10) >DroEre_CAF1 25953 112 - 1 GGUACUCAUUAUGCCACCGAAAUGAAUGCAAAUCGGCGGCAUUAACUUCACAUCCUCCGGCACAAUUAAGCGCCCU-UCUGCCGU-CC------CACUCGCACAUCCGACGGGUGCAUCU ((((......(((((.((((..((....))..)))).)))))................(((.(......).)))..-..))))..-..------(((((((......).))))))..... ( -28.20) >consensus GCGACUCAUUAUGCCACCGAAAUGAAUGCAAAUCGGCGGCAUUAACUUCACAUCCUCCGGUACAAUUAAGUGCCCU_UCUGCCGU_CC______CACUCGCAC______CGAGUGGAUCU ..........(((((.((((..((....))..)))).)))))................(((((......)))))....................((((((.........))))))..... (-22.60 = -22.44 + -0.16)



| Location | 20,588,068 – 20,588,187 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.35 |
| Mean single sequence MFE | -41.04 |
| Consensus MFE | -27.04 |
| Energy contribution | -29.84 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20588068 119 + 23771897 AGA-AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGCGGUCCCCAACUGCAAAGAGGUCGAGGUCAAUCUGCGAAGA (((-..(((((((..(((((..((.((((((((.((((.(((((((((..((....))..)))))))))))))...)))).))))))...)))))..))))....)))..)))....... ( -40.30) >DroSec_CAF1 32847 119 + 1 AGA-AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGCGGUCCCCCACUGCAAAGAGGUCGAGGUCAAUCUGCCAAGA ...-..((((....((((.(((((.((((((((.((((.(((((((((..((....))..)))))))))))))...)))).)))).)).((....)).......))))))).)))).... ( -43.00) >DroAna_CAF1 54367 110 + 1 AGACAGGGCGA---ACAGC----AAGGAUGUGACGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUAGCAUAAUGAGCUCAGGC---CCACUGCAAAGAGGUCGAGGUCAAGCUGAGAGGA ...........---.((((----...............((((..((((..((....))..))))..))))..(((.(((.(((---(..((....)).))))))).))).))))...... ( -32.30) >DroSim_CAF1 26348 119 + 1 AGA-AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGCGGUCCCCAACUGCAAAGAGGUCGAGGUCAAUCUGCGAAGA (((-..(((((((..(((((..((.((((((((.((((.(((((((((..((....))..)))))))))))))...)))).))))))...)))))..))))....)))..)))....... ( -40.30) >DroEre_CAF1 25986 119 + 1 AGA-AGGGCGCUUAAUUGUGCCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUACCCGUCCCCAGCUGCAAGGAGGUCGAGGUCAAUUUGCGGCGA ...-..(((((......)))))((.(((((.(.(.(((((((((((((..((....))..))))))))))..))).).).))))))).((((((((..((......))..)))))))).. ( -49.30) >consensus AGA_AGGGCACUUAAUUGUACCGGAGGAUGUGAAGUUAAUGCCGCCGAUUUGCAUUCAUUUCGGUGGCAUAAUGAGUCGCGGUCCCCAACUGCAAAGAGGUCGAGGUCAAUCUGCGAAGA (((...(((((((..(((((..((.((((((((.((((.(((((((((..((....))..)))))))))))))...)))).))))))...)))))..))))....)))..)))....... (-27.04 = -29.84 + 2.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:53 2006