
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,583,598 – 20,583,753 |
| Length | 155 |
| Max. P | 0.839234 |

| Location | 20,583,598 – 20,583,716 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -33.46 |
| Energy contribution | -33.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
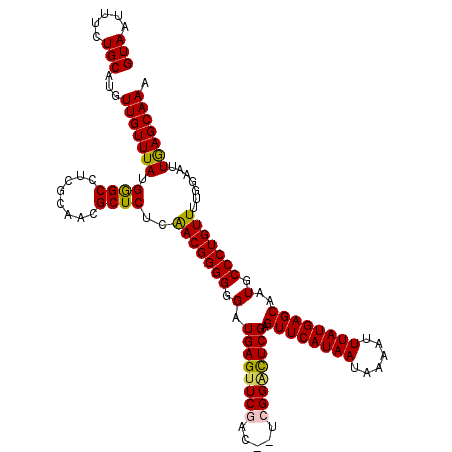
>3L_DroMel_CAF1 20583598 118 + 23771897 GUAAUUUCUGCAUGUUGUUUAUGGGCCUCGCAACGCUCUCAACGGGGGGAUGAGUUCGAC--UCGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAA .(((((((.((.(((((.....((((........)))).)))))((((.(((((((((..--.)))))))..((((((((......)))))))).)).))))))...)))))))...... ( -35.40) >DroSec_CAF1 28313 118 + 1 GUAAUUUCUGCAUGUUGUUUAUGGGCCUCGCAACGCUCUCAACGGGGGGAUGAGUUCGAC--UGGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAA .(((((((.((.(((((.....((((........)))).)))))((((.((((((((...--..))))))..((((((((......)))))))).)).))))))...)))))))...... ( -34.70) >DroAna_CAF1 49735 120 + 1 GUAAUUUCUGCAUGUUGUUUAUGAGCCUCGCAACGCUCUCGACGGGGGGAUGAGUUCGGCUCUCGGGUUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAA .(((((((.(((..........(((((..((....(((((....)))))....))..)))))..((((....((((((((......))))))))...)))))))...)))))))...... ( -34.70) >DroSim_CAF1 21847 118 + 1 GUAAUUUCUGCAUGUUGUUUAUGGGCCUCGCAACGCUCUCAACGGGGGGAUGAGUUCGAC--UGGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAA .(((((((.((.(((((.....((((........)))).)))))((((.((((((((...--..))))))..((((((((......)))))))).)).))))))...)))))))...... ( -34.70) >DroEre_CAF1 21441 118 + 1 GUAAUUUCUGCAUGUUGUUUAUGGGCCUCGCAACGCUCUCAACGGGGGGAUGAGUUCGCC--UCGGCCUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAAUAAGCAAA (((.....)))...((((((((((((........))))..(((((((.(.((((..((..--.))..)))).((((((((......))))))))..).)))))))......)))))))). ( -32.70) >consensus GUAAUUUCUGCAUGUUGUUUAUGGGCCUCGCAACGCUCUCAACGGGGGGAUGAGUUCGAC__UCGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAA (((.....)))...(((((((.((((........))))..(((((((.(.((((((((.....)))))))).((((((((......))))))))..).))))))).......))))))). (-33.46 = -33.30 + -0.16)

| Location | 20,583,638 – 20,583,753 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.33 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20583638 115 + 23771897 AACGGGGGGAUGAGUUCGAC--UCGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAAUCAAAAAAGGAAGGGGCCCACUGAG---GAGGCACAUGUG ..(((.((..((((((((..--.)))))))).((((((((......))))))))...(((((.((((....((((.....))))...)))).))))))).)))..---............ ( -32.80) >DroSec_CAF1 28353 118 + 1 AACGGGGGGAUGAGUUCGAC--UGGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAAUCAAAAAAGGAAGGGGCCCACUGCGGCGGAGUCACAUGUG ..((.((((.(((((((...--..))))))).((((((((......))))))))...(((((.((((....((((.....))))...)))).))))))).)).))............... ( -34.00) >DroAna_CAF1 49775 116 + 1 GACGGGGGGAUGAGUUCGGCUCUCGGGUUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAAUCAAAAGACCUGGAUAUA-AAA---AAGGAGUCACAUGUG (((.((((.(((((..((.....))..)))..((((((((......)))))))).)).))))(((..((..((((.....))))....))..)))...-...---.....)))....... ( -28.60) >DroSim_CAF1 21887 117 + 1 AACGGGGGGAUGAGUUCGAC--UGGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAAUCAAA-AAGGAAGGGGCCCACUGCGGCGGAGUCACAUGUG ..((.((((.(((((((...--..))))))).((((((((......))))))))...(((((.((((....((((.....)))).-.)))).))))))).)).))............... ( -33.70) >DroEre_CAF1 21481 114 + 1 AACGGGGGGAUGAGUUCGCC--UCGGCCUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAAUAAGCAAAUCAAAAAAGGAAGGGGCC-ACA---AAGGAGGCACAUGUG ..(((((.((.....)).))--)))(((((..((((((((......))))))))...(((((.((((((...........))))))......))))).-...---...)))))....... ( -37.70) >consensus AACGGGGGGAUGAGUUCGAC__UCGGACUCGAGUUCAUAAUAAAAUUUAUGAGCAAUGCCCUGUUUUGGAAUUGAGCAAAUCAAAAAAGGAAGGGGCCCACUG_GAAGGAGUCACAUGUG (((((((.(.((((((((.....)))))))).((((((((......))))))))..).)))))))....................................................... (-24.46 = -24.62 + 0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:40 2006