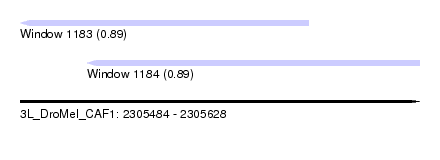
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,305,484 – 2,305,628 |
| Length | 144 |
| Max. P | 0.894972 |

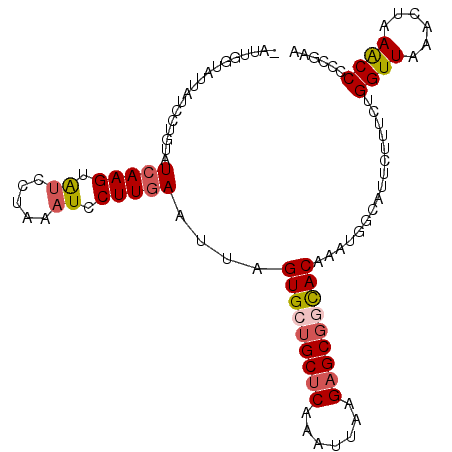
| Location | 2,305,484 – 2,305,588 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.06 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2305484 104 - 23771897 AAUUGGUAUUAUCCUGUAUCAAGUAUCCUGAAUCCUUGAAUUCGUGCUGCUCAAAUUAAGAGCGGUACAAAUGGCAUUCUUUCUGGUUAAACUUAACCCCCGAA ..(((((((......))))))).......((((((....(((.(((((((((.......))))))))).))))).)))).....(((((....)))))...... ( -24.80) >DroSec_CAF1 54703 104 - 1 UAUUGGUAUUAUCCUGUAUCAAGUAUCCUAAAUCCUUGAAUUAGUGCUGCUCAAAUUAAGAGCGGCACAAAUGGCAUUCUUUCUGGUUAAACUAAACCCCCGAA ..((((........(((.(((((.((.....)).)))))....(((((((((.......))))))))).....)))........((((......)))).)))). ( -25.30) >DroSim_CAF1 54644 104 - 1 UAUUGGUAUUAUCCUCUAUCAAGUAGCCUAAAUCCUUGAAUUAGUGCUGCUCAAAUUAAGAGCGGCACAAAUGGCAUUCUUUCUGGUUAAACUAAGCCCCCGAA ..((((((........)))))).(((((..((....((.(((.(((((((((.......))))))))).)))..))....))..)))))............... ( -25.10) >DroEre_CAF1 57903 92 - 1 ------------CAUAUCUGAAGUGUCCCAAAUCCUUGAAUUAGUGUGGCGCAAAUUAAGAGCGUCACAAAUGGCAUUUUCUCUGGUUAAACUAAACCCCAGAA ------------....(((((((((((.(((....)))......((((((((.........))))))))...))))))).....((((......)))).)))). ( -22.60) >DroYak_CAF1 57407 95 - 1 -AUU--------UAUGUUUCAAGUGUACUAAAUCCUUGAAUUAGUGUUGCUCAAAUUAAGAGCGUCACAAAUGGCAUUCUCUCUGGUUGAACUAAACCUCAGAA -...--------.((((((((((.(........)))))))...(((.(((((.......))))).))).....))))..(((..((((......))))..))). ( -21.10) >consensus _AUUGGUAUUAUCCUGUAUCAAGUAUCCUAAAUCCUUGAAUUAGUGCUGCUCAAAUUAAGAGCGGCACAAAUGGCAUUCUUUCUGGUUAAACUAAACCCCCGAA ..................(((((.((.....)).)))))....(((((((((.......)))))))))................((((......))))...... (-16.42 = -17.06 + 0.64)

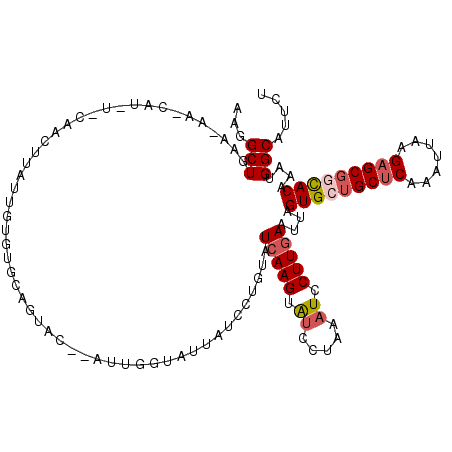
| Location | 2,305,508 – 2,305,628 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2305508 120 - 23771897 AAGGCUGAAUAACCAUUUCCAACUCAUUGUGUUCAGUACGAAUUGGUAUUAUCCUGUAUCAAGUAUCCUGAAUCCUUGAAUUCGUGCUGCUCAAAUUAAGAGCGGUACAAAUGGCAUUCU ......((((..((((((.....(((..(..(((((......(((((((......))))))).....)))))..).)))....(((((((((.......))))))))))))))).)))). ( -33.00) >DroSec_CAF1 54727 110 - 1 AAGGCUGAA----------CAACUUAUUGUGUGCAGUACAUAUUGGUAUUAUCCUGUAUCAAGUAUCCUAAAUCCUUGAAUUAGUGCUGCUCAAAUUAAGAGCGGCACAAAUGGCAUUCU ...((((.(----------(((....))))...)))).(((.(((((((......))))))).....................(((((((((.......)))))))))..)))....... ( -27.90) >DroSim_CAF1 54668 110 - 1 AAGGCUGAA----------CAACUUAUUGUGUGCAGUACAUAUUGGUAUUAUCCUCUAUCAAGUAGCCUAAAUCCUUGAAUUAGUGCUGCUCAAAUUAAGAGCGGCACAAAUGGCAUUCU .((((((..----------.....(((((....)))))....((((((........)))))).)))))).......((.(((.(((((((((.......))))))))).)))..)).... ( -29.80) >DroEre_CAF1 57927 106 - 1 AAGGCUGAACAACCAUUUGAAACUUAUUGUGUGUAUAA--------------CAUAUCUGAAGUGUCCCAAAUCCUUGAAUUAGUGUGGCGCAAAUUAAGAGCGUCACAAAUGGCAUUUU ...(((...(((..(((((..((((..(((((.....)--------------))))....))))....)))))..)))......((((((((.........))))))))...)))..... ( -24.40) >DroYak_CAF1 57431 109 - 1 AAGGCUGAACAAUCAUAUCAAACUUAUUAGGUAUAUAU---AUU--------UAUGUUUCAAGUGUACUAAAUCCUUGAAUUAGUGUUGCUCAAAUUAAGAGCGUCACAAAUGGCAUUCU ...(((...........((((....(((.(((((((..---...--------..........))))))).)))..))))....(((.(((((.......))))).)))....)))..... ( -17.86) >consensus AAGGCUGAA_AA_CAU_U_CAACUUAUUGUGUGCAGUAC__AUUGGUAUUAUCCUGUAUCAAGUAUCCUAAAUCCUUGAAUUAGUGCUGCUCAAAUUAAGAGCGGCACAAAUGGCAUUCU ...(((....................................................(((((.((.....)).)))))....(((((((((.......)))))))))....)))..... (-16.70 = -17.50 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:45 2006