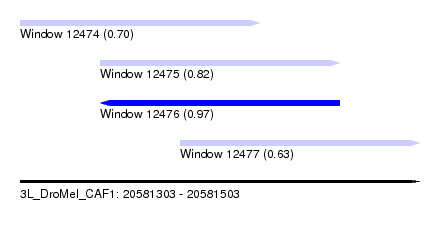
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,581,303 – 20,581,503 |
| Length | 200 |
| Max. P | 0.969398 |

| Location | 20,581,303 – 20,581,423 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -33.86 |
| Energy contribution | -33.86 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20581303 120 + 23771897 GUUCGUGGCCGUUUCUGUGCUGUUAGAUGAAGCGCGAGUUUGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGG ..(((((.(((..((..(((((((((.(.((((....)))).)))))).....(((((((((((.....)))).....)))))))......))))...)).))))))))........... ( -39.40) >DroSec_CAF1 25995 120 + 1 GUUCGUGGCCGUUUCUGUGCUGUUAGAUGAAGCGCGAGUUUGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGG ..(((((.(((..((..(((((((((.(.((((....)))).)))))).....(((((((((((.....)))).....)))))))......))))...)).))))))))........... ( -39.40) >DroAna_CAF1 47540 109 + 1 GUUCGU------UU-----CUGUUAGAUGAAGCGCGAGUUUGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCCGAGCGAUGAUGAACCGG ((((((------(.-----..(((((.(.((((....)))).))))))..((((((((((((((.....)))).....))))))).((((........))))...)))..)))))))... ( -33.90) >DroSim_CAF1 19556 120 + 1 GUUCGUGGCCGUUUCUGUGCUGUUAGAUGAAGCGCGAGUUUGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGG ..(((((.(((..((..(((((((((.(.((((....)))).)))))).....(((((((((((.....)))).....)))))))......))))...)).))))))))........... ( -39.40) >DroEre_CAF1 19183 120 + 1 GUUCGUGGCCGUUUCUGUGCUGUUAGAUGAAGCGCGAGUUUGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCCGAGCGAUGAUGAACCGG ..((((.((((..((..(((((((((.(.((((....)))).)))))).....(((((((((((.....)))).....)))))))......))))...))..)).)).))))........ ( -39.10) >consensus GUUCGUGGCCGUUUCUGUGCUGUUAGAUGAAGCGCGAGUUUGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGG ((((((...............(((((.(.((((....)))).))))))..((((((((((((((.....)))).....))))))).((((........))))...)))...))))))... (-33.86 = -33.86 + -0.00)

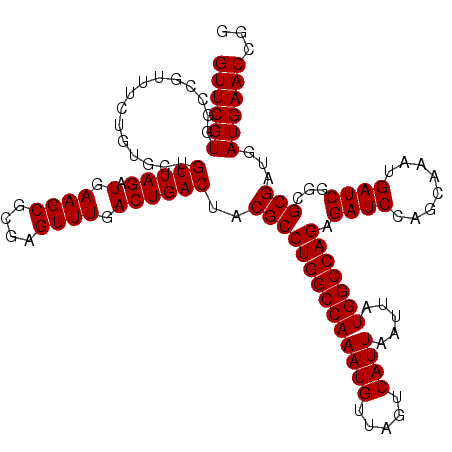
| Location | 20,581,343 – 20,581,463 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20581343 120 + 23771897 UGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGAACCUCCCCGAAGU ...(((.....(((((((((((((.....)))).....))))))..((((........)))))))((((((((((...(....))))))))))).........))).............. ( -34.00) >DroSec_CAF1 26035 120 + 1 UGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGACCCUCCCCGAAGU ...(((.....(((((((((((((.....)))).....))))))..((((........)))))))((((((((((...(....))))))))))).........))).............. ( -34.00) >DroAna_CAF1 47569 119 + 1 UGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCCGAGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGUCGC-CCCUUUGGU .(((((....((.(((((((((((.....)))).....))))))).((((........)))))).((((((((((...(....))))))))))).........)))))((-(.....))) ( -36.20) >DroSim_CAF1 19596 120 + 1 UGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGACCCUCCCCGAAGU ...(((.....(((((((((((((.....)))).....))))))..((((........)))))))((((((((((...(....))))))))))).........))).............. ( -34.00) >DroEre_CAF1 19223 118 + 1 UGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCCGAGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGAC-CUCCCCG-AGU ......(((.((.(((((((((((.....)))).....))))))).((((........)))).(((.((((((((...(....)))))))))((((....))))....-)))..))-))) ( -31.80) >consensus UGACUGACUACGCCUGGCCAAAUGUUAGUCAUUAAUUAUGGCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGACCCUCCCCGAAGU ...........(((((((((((((.....)))).....))))))).((((........))))...))((((((((...(....)))))))))((((....))))................ (-31.60 = -31.60 + -0.00)


| Location | 20,581,343 – 20,581,463 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -34.62 |
| Energy contribution | -35.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20581343 120 - 23771897 ACUUCGGGGAGGUUCUGUCAAUAAUGACGAUGAUGAGUCGCCGGUUCAUCAUCGCGCCGAUCAUUUGCUGGAUCUCUGGCCAUAAUUAAUGACUAACAUUUGGCCAGGCGUAGUCAGUCA .(((....))).............((((((((((((..(...)..))))))))(((((((((........))))..((((((.....((((.....))))))))))))))).)))).... ( -39.70) >DroSec_CAF1 26035 120 - 1 ACUUCGGGGAGGGUCUGUCAAUAAUGACGAUGAUGAGUCGCCGGUUCAUCAUCGCGCCGAUCAUUUGCUGGAUCUCUGGCCAUAAUUAAUGACUAACAUUUGGCCAGGCGUAGUCAGUCA .(....).((..(.((((((....))))((((((((..(...)..))))))))(((((((((........))))..((((((.....((((.....))))))))))))))))).)..)). ( -39.90) >DroAna_CAF1 47569 119 - 1 ACCAAAGGG-GCGACUGUCAAUAAUGACGAUGAUGAGUCGCCGGUUCAUCAUCGCUCGGAUCAUUUGCUGGAUCUCUGGCCAUAAUUAAUGACUAACAUUUGGCCAGGCGUAGUCAGUCA .((....))-..(((((.(......(((((((((((..(...)..))))))))).))(((((........)))))(((((((.....((((.....))))))))))).....).))))). ( -40.20) >DroSim_CAF1 19596 120 - 1 ACUUCGGGGAGGGUCUGUCAAUAAUGACGAUGAUGAGUCGCCGGUUCAUCAUCGCGCCGAUCAUUUGCUGGAUCUCUGGCCAUAAUUAAUGACUAACAUUUGGCCAGGCGUAGUCAGUCA .(....).((..(.((((((....))))((((((((..(...)..))))))))(((((((((........))))..((((((.....((((.....))))))))))))))))).)..)). ( -39.90) >DroEre_CAF1 19223 118 - 1 ACU-CGGGGAG-GUCUGUCAAUAAUGACGAUGAUGAGUCGCCGGUUCAUCAUCGCUCGGAUCAUUUGCUGGAUCUCUGGCCAUAAUUAAUGACUAACAUUUGGCCAGGCGUAGUCAGUCA ...-...((((-(((((.(((...((((((((((((..(...)..))))))))).)))......))).)))))))))(((((.....((((.....))))))))).(((.......))). ( -37.90) >consensus ACUUCGGGGAGGGUCUGUCAAUAAUGACGAUGAUGAGUCGCCGGUUCAUCAUCGCGCCGAUCAUUUGCUGGAUCUCUGGCCAUAAUUAAUGACUAACAUUUGGCCAGGCGUAGUCAGUCA ........................((((((((((((..(...)..))))))))((((.((((........)))).(((((((.....((((.....))))))))))))))).)))).... (-34.62 = -35.02 + 0.40)

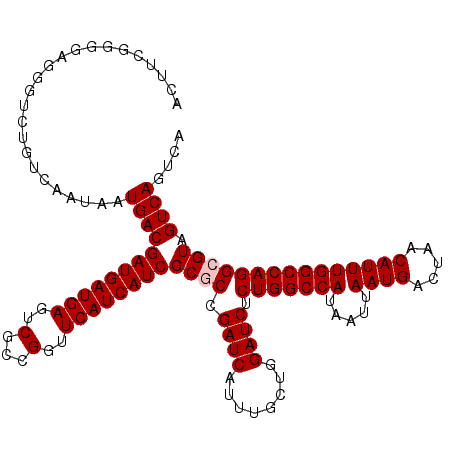
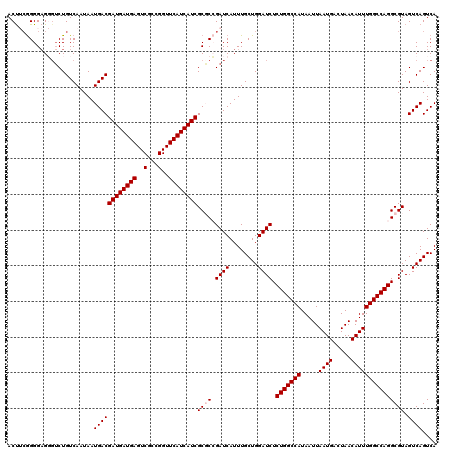
| Location | 20,581,383 – 20,581,503 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.74 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20581383 120 + 23771897 GCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGAACCUCCCCGAAGUCCGCAGACCCCGCGAAACAUGUUCUUUAAACUUUGGCCUU (((((((.....((......((((...((((((((...(....)))))))))((((....)))).........)))).(((....)))...))(((.....)))......)))))))... ( -33.00) >DroSec_CAF1 26075 120 + 1 GCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGACCCUCCCCGAAGUCCGCGGACCCCGCGAAACAUGUUCUUUAAACUUUGGCCUU (((((((....((...(((.((((...((((((((...(....)))))))))((((....)))).........))))....(((((...)))))...)))...)).....)))))))... ( -37.10) >DroAna_CAF1 47609 118 + 1 GCCAGAGAUCCAGCAAAUGAUCCGAGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGUCGC-CCCUUUGGUCCUCCUCCCCCUCC-AGUGUGUUCUUUAAACUUUGGCCUU (((((((....((((..((....(((.((.(((.((..(((((((.......((((....))))))))))-)...)).))).)))))......)-)...)))).......)))))))... ( -31.41) >DroSim_CAF1 19636 120 + 1 GCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGACCCUCCCCGAAGUCCGCGGACCCCGCGAAACAUGUUCUUUAAACUUUGGCCUU (((((((....((...(((.((((...((((((((...(....)))))))))((((....)))).........))))....(((((...)))))...)))...)).....)))))))... ( -37.10) >DroEre_CAF1 19263 118 + 1 GCCAGAGAUCCAGCAAAUGAUCCGAGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGAC-CUCCCCG-AGUCCGCGGACCCCGCGAAACAUGUUCUUUAAACUUUGGCCUU (((((((...((.....))....((((((((((((...(....)))))))))((((....)))).(((-.......-.)))(((((...)))))......))))......)))))))... ( -35.50) >consensus GCCAGAGAUCCAGCAAAUGAUCGGCGCGAUGAUGAACCGGCGACUCAUCAUCGUCAUUAUUGACAGACCCUCCCCGAAGUCCGCGGACCCCGCGAAACAUGUUCUUUAAACUUUGGCCUU (((((((.....(..((((((....((((((((((...(....))))))))))).))))))..).................((((.....))))................)))))))... (-27.74 = -28.74 + 1.00)

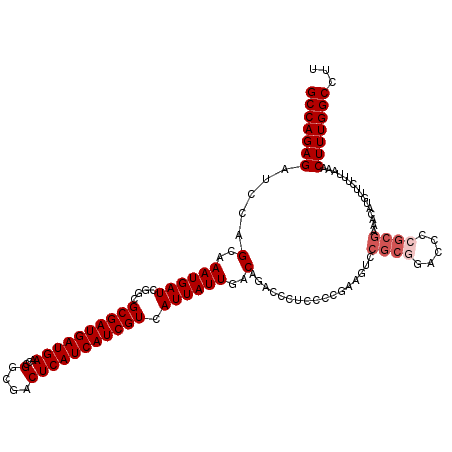

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:35 2006