
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
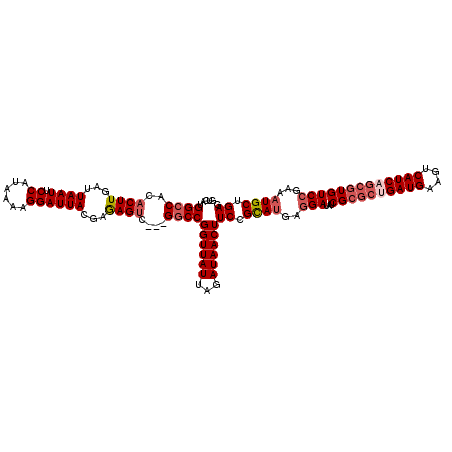
| Location | 20,577,453 – 20,577,570 |
| Length | 117 |
| Max. P | 0.993782 |

| Location | 20,577,453 – 20,577,570 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -31.46 |
| Energy contribution | -32.94 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
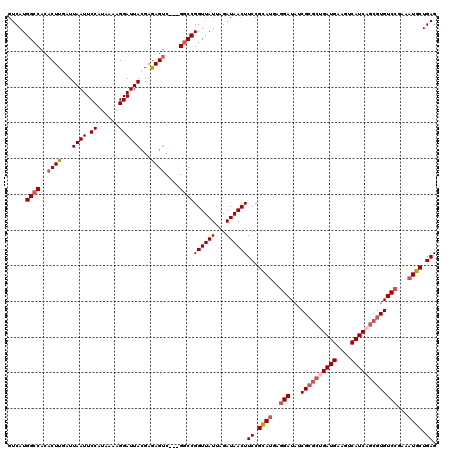
>3L_DroMel_CAF1 20577453 117 + 23771897 GUCAUGGCCACACUUGAUUAAUUCCAUAAAAGGAUUACGAAAGUC---GGCCGGUUAUUAGAUAACUUCCGCAUGAGGAUAUCGCGCUGAUGAAGUCAUCAGCGUGUCCGAAAUGCUGAG .....((((..((((...((((.((......))))))...)))).---))))((((((...))))))((.((((..(((...((((((((((....)))))))))))))...)))).)). ( -40.50) >DroSec_CAF1 22209 117 + 1 GUCAUGGCCACACUUAAUUAAUUCCAUAAAAGGAUUACGAGAGUC---GGCCGGUUAUUAGAUAACUUCCGCAUGAGGAUAUCGCGCUGAUGAAGUCAUCAGCGUGUCCGAAAUGCUGAG .....((((((.(((...((((.((......)))))).))).)).---))))((((((...))))))((.((((..(((...((((((((((....)))))))))))))...)))).)). ( -40.00) >DroAna_CAF1 43853 115 + 1 GUCAUGGCCACACUUGAUUAAUUCCAUAAAAGGAUUAUGAGAGUCGUCGCCCGGUUAUUAGAUAACUUCCGUAUAGCGAU--CG---AGAUGAAGUCAUCAGCGUGUCCGAAAUGCUGAG ....((((..(((((((((...(((......))).((((..(((..((............))..)))..))))....)))--))---)).))..))))(((((((.......))))))). ( -29.90) >DroSim_CAF1 15744 117 + 1 GUCAUGGCCACACUUGAUUAAUUCCAUAAAAGGAUUACGAGAGUC---GGCCGGUUAUUAGAUAACUUCCGCAUGAGGAUAUCGCGCUGAUGAAGUCAUCAGCGUGUCCGAAAUGCUGAG .....((((((.((((..((((.((......)))))))))).)).---))))((((((...))))))((.((((..(((...((((((((((....)))))))))))))...)))).)). ( -42.20) >DroEre_CAF1 15554 117 + 1 GUCAUGGCCACACUUGAUUAAUUCCAUAAAAGGAUUACGAGAGCC---GGCCGGUUAUUAGAUAACUUCCGCAGGAGGAUAUCGCGCCGAUGAAGUCAUCAGCGUGUCCGAAAUGCUGAG .((((((((.(.((((..((((.((......)))))))))).)..---))))(((.....((((.(((((...))))).))))..))).)))).....(((((((.......))))))). ( -34.30) >consensus GUCAUGGCCACACUUGAUUAAUUCCAUAAAAGGAUUACGAGAGUC___GGCCGGUUAUUAGAUAACUUCCGCAUGAGGAUAUCGCGCUGAUGAAGUCAUCAGCGUGUCCGAAAUGCUGAG .....((((..((((...((((.((......))))))...))))....))))((((((...))))))((.((((..(((...((((((((((....)))))))))))))...)))).)). (-31.46 = -32.94 + 1.48)

| Location | 20,577,453 – 20,577,570 |
|---|---|
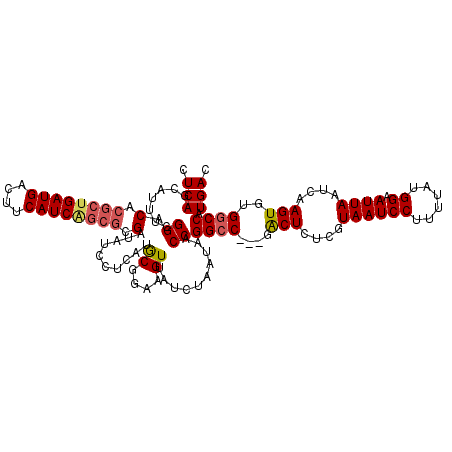
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.32 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.48 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20577453 117 - 23771897 CUCAGCAUUUCGGACACGCUGAUGACUUCAUCAGCGCGAUAUCCUCAUGCGGAAGUUAUCUAAUAACCGGCC---GACUUUCGUAAUCCUUUUAUGGAAUUAAUCAAGUGUGGCCAUGAC .((.((((...((((.((((((((....)))))))).)...)))..)))).)).(((((...))))).((((---(((((...((((((......)).))))...)))).)))))..... ( -36.40) >DroSec_CAF1 22209 117 - 1 CUCAGCAUUUCGGACACGCUGAUGACUUCAUCAGCGCGAUAUCCUCAUGCGGAAGUUAUCUAAUAACCGGCC---GACUCUCGUAAUCCUUUUAUGGAAUUAAUUAAGUGUGGCCAUGAC .((.((((...((((.((((((((....)))))))).)...)))..)))).)).(((((...))))).((((---((((....((((((......)).))))....))).)))))..... ( -35.20) >DroAna_CAF1 43853 115 - 1 CUCAGCAUUUCGGACACGCUGAUGACUUCAUCU---CG--AUCGCUAUACGGAAGUUAUCUAAUAACCGGGCGACGACUCUCAUAAUCCUUUUAUGGAAUUAAUCAAGUGUGGCCAUGAC ...........((.(((((((((...(((...(---((--.(((((...(((..(((....)))..)))))))))))....(((((.....))))))))...))).)))))).))..... ( -28.10) >DroSim_CAF1 15744 117 - 1 CUCAGCAUUUCGGACACGCUGAUGACUUCAUCAGCGCGAUAUCCUCAUGCGGAAGUUAUCUAAUAACCGGCC---GACUCUCGUAAUCCUUUUAUGGAAUUAAUCAAGUGUGGCCAUGAC .((.((((...((((.((((((((....)))))))).)...)))..)))).)).(((((...))))).((((---((((....((((((......)).))))....))).)))))..... ( -35.20) >DroEre_CAF1 15554 117 - 1 CUCAGCAUUUCGGACACGCUGAUGACUUCAUCGGCGCGAUAUCCUCCUGCGGAAGUUAUCUAAUAACCGGCC---GGCUCUCGUAAUCCUUUUAUGGAAUUAAUCAAGUGUGGCCAUGAC .(((.......((...((((((((....)))))))).((((.(.(((...))).).))))......))((((---((((....((((((......)).))))....))).))))).))). ( -33.30) >consensus CUCAGCAUUUCGGACACGCUGAUGACUUCAUCAGCGCGAUAUCCUCAUGCGGAAGUUAUCUAAUAACCGGCC___GACUCUCGUAAUCCUUUUAUGGAAUUAAUCAAGUGUGGCCAUGAC .(((.......((.(.((((((((....)))))))).)..........((....))..........))((((....(((....((((((......)).))))....)))..)))).))). (-25.92 = -26.48 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:20 2006