
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,575,600 – 20,575,697 |
| Length | 97 |
| Max. P | 0.995681 |

| Location | 20,575,600 – 20,575,697 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -24.49 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
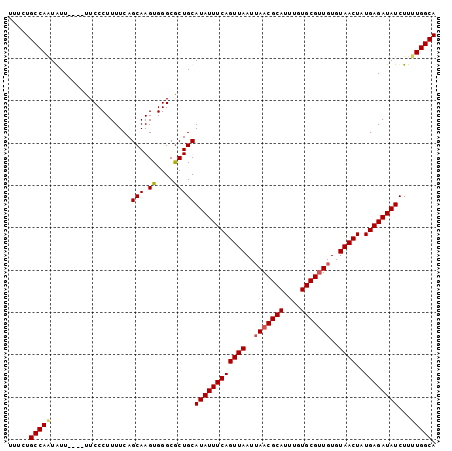
>3L_DroMel_CAF1 20575600 97 + 23771897 UUUCUGCCAAUGUU----UUCCCUUUUCAGCAAGUGGGCACUGCAUAUUUCAGUUAAUUAACGCAUUUGUGCGUUGUGUAACUAUGAGAUAUCUUAUGGCA .....((((.(((.----..(((............)))....)))(((((((((((..(((((((....)))))))..))))..))))))).....)))). ( -25.80) >DroSec_CAF1 20383 97 + 1 UUUCUGCCAAUAUU----UUCCCUUUUCAGCAAGUGGGCGCUGCAUAUUUCAGUUAAUAAACGCAUUUGUGCGUUGUGUAACUAUGAGAUAUCUUUUGGCA .....(((((....----.........((((........)))).((((((((((((...((((((....))))))...))))..))))))))...))))). ( -26.80) >DroSim_CAF1 13892 97 + 1 UUUCUGCCAAUAUU----UUCCCUUUUCAGCAAGUGGGCGCUGCAUAUUUCAGUUAAUUAACGCAUUUGUGCGUUGUGUAACUAUGAGAUAUCUUUUGGCA .....(((((....----.........((((........)))).((((((((((((..(((((((....)))))))..))))..))))))))...))))). ( -26.80) >DroEre_CAF1 13674 101 + 1 UUUCUGCCAGUGUUUUGUUUCCCUUUUCAGCAAGUGGGCGCUGCAUAUUUCAGUUAAUUAACGCACUUGUGCGCUGUGUAACUAUGAGAUAUCUUUUGGCA .....((((((((...((.((((((......))).))).)).)))(((((((((((.....((((....)))).....))))..)))))))....))))). ( -24.50) >consensus UUUCUGCCAAUAUU____UUCCCUUUUCAGCAAGUGGGCGCUGCAUAUUUCAGUUAAUUAACGCAUUUGUGCGUUGUGUAACUAUGAGAUAUCUUUUGGCA .....(((((...................(((.((....)))))((((((((((((..(((((((....)))))))..))))..))))))))...))))). (-24.49 = -24.92 + 0.44)


| Location | 20,575,600 – 20,575,697 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.71 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
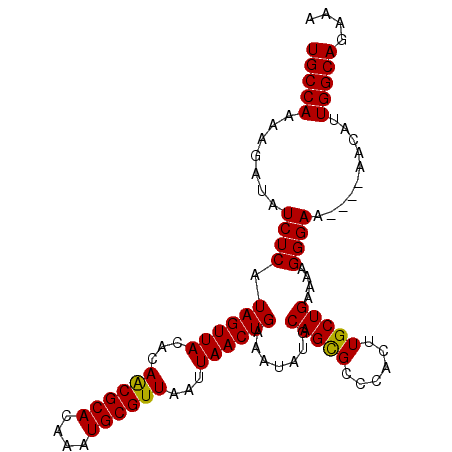
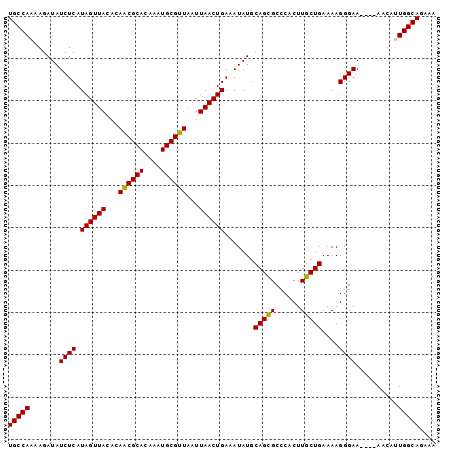
>3L_DroMel_CAF1 20575600 97 - 23771897 UGCCAUAAGAUAUCUCAUAGUUACACAACGCACAAAUGCGUUAAUUAACUGAAAUAUGCAGUGCCCACUUGCUGAAAAGGGAA----AACAUUGGCAGAAA (((((.......((((.((((((...((((((....))))))...)))))).......((((........))))....)))).----.....))))).... ( -23.82) >DroSec_CAF1 20383 97 - 1 UGCCAAAAGAUAUCUCAUAGUUACACAACGCACAAAUGCGUUUAUUAACUGAAAUAUGCAGCGCCCACUUGCUGAAAAGGGAA----AAUAUUGGCAGAAA ((((((......((((.((((((...((((((....))))))...)))))).......(((((......)))))....)))).----....)))))).... ( -26.20) >DroSim_CAF1 13892 97 - 1 UGCCAAAAGAUAUCUCAUAGUUACACAACGCACAAAUGCGUUAAUUAACUGAAAUAUGCAGCGCCCACUUGCUGAAAAGGGAA----AAUAUUGGCAGAAA ((((((......((((.((((((...((((((....))))))...)))))).......(((((......)))))....)))).----....)))))).... ( -26.20) >DroEre_CAF1 13674 101 - 1 UGCCAAAAGAUAUCUCAUAGUUACACAGCGCACAAGUGCGUUAAUUAACUGAAAUAUGCAGCGCCCACUUGCUGAAAAGGGAAACAAAACACUGGCAGAAA (((((.......((((.((((((...((((((....))))))...)))))).......(((((......)))))....))))..........))))).... ( -25.23) >consensus UGCCAAAAGAUAUCUCAUAGUUACACAACGCACAAAUGCGUUAAUUAACUGAAAUAUGCAGCGCCCACUUGCUGAAAAGGGAA____AACAUUGGCAGAAA (((((.......((((.((((((...((((((....))))))...)))))).......(((((......)))))....))))..........))))).... (-24.08 = -23.71 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:16 2006