
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,573,147 – 20,573,316 |
| Length | 169 |
| Max. P | 0.633825 |

| Location | 20,573,147 – 20,573,256 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -28.01 |
| Energy contribution | -27.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
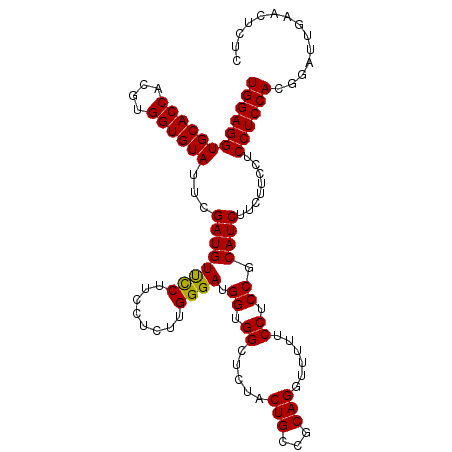
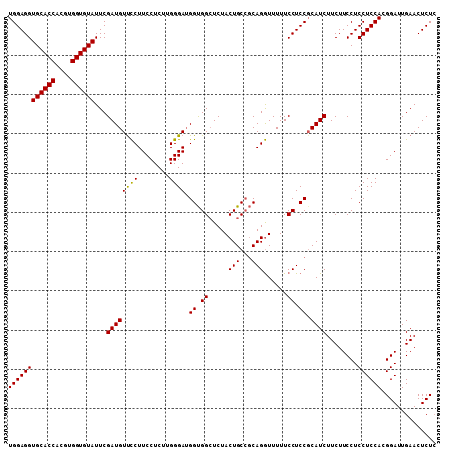
>3L_DroMel_CAF1 20573147 109 - 23771897 UGGAGGUGCACCACGUGGUGUAUUCGAUGUCUCUUCCUCUUGGGAUGGCGGCUCUACUGCCCCAGAUUUUUCCUCCGCAUCUUCUUCCUCCUCCACGGAUUGAACUCUA ((((((((((((....))))))...(((((...((((....))))(((.(((......))))))............)))))........)))))).............. ( -33.30) >DroSec_CAF1 18063 109 - 1 UGGAGGUGCACCACGUGGUGUAUUCGAUGUUCCUUCCUCUUGGGAUGGUGGCUCUACUGCCGCAGGUUUUUCCUCCUCAUCUUCUUCCUCCUCCAUGGAUUGAACUCUC ..(((.((((((....)))))))))((.((((..(((....((((.((((((......)))))(((.....)))............).))))....)))..)))))).. ( -33.30) >DroSim_CAF1 11560 109 - 1 UGGAGGUGCACCACGUGGUGUAUUCGAUGUUCCUUCCUCUUGGGAAGGUGGAUCUACUGCAGCAGGUUUUUCCUCCGCAUCUUCUUCCUCCUCCACGGAUUGAACUCUC ((((((((((((....))))))...(((((.((((((.....)))))).(((...(((......)))...)))...)))))........)))))).............. ( -35.30) >consensus UGGAGGUGCACCACGUGGUGUAUUCGAUGUUCCUUCCUCUUGGGAUGGUGGCUCUACUGCCGCAGGUUUUUCCUCCGCAUCUUCUUCCUCCUCCACGGAUUGAACUCUC ((((((((((((....))))))...((((((((........)))).((.((.....(((...)))......)).)).))))........)))))).............. (-28.01 = -27.57 + -0.44)

| Location | 20,573,226 – 20,573,316 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -33.11 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.41 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
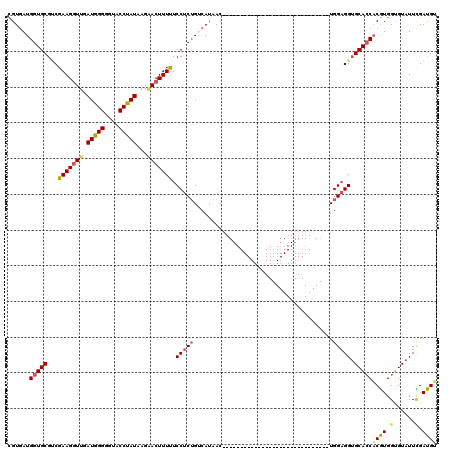
>3L_DroMel_CAF1 20573226 90 - 23771897 CGUGAUGGUGCGCCGAAGGUUGAUGGGGGUACCUAUAAGAACUUUUUCCUCUGACAUAAC------------------------------UGGAGGUGCACCACGUGGUGUAUUCGAUGU ......(((((((((((((((.(((((....)))))...)))))))(((...........------------------------------.)))))))))))((((.(......).)))) ( -29.80) >DroSec_CAF1 18142 90 - 1 CGUGAUGGUGCGUCGAAGGUUGAUAGGGGUACCUAUAAGAACUUUUUCCUCUGUCAUAAC------------------------------UGGAGGUGCACCACGUGGUGUAUUCGAUGU .........(((((((((..(((((((((..................)))))))))...)------------------------------)....(((((((....)))))))))))))) ( -28.47) >DroAna_CAF1 39792 120 - 1 GGUGAUGGUGCGGCGAAGAUUGAUGGGCGUGCCUAUCAGGACUUUCUCCUCGGACACCACAGUGGGCUGGGAUUGUGGCUGGGGCUGUGUUGGUGGAGCAGCCCUUGGGGUGUUGGAGGG ..((((((..((.(.(.......).).))..)).)))).........((((.((((((.((...((((........))))((((((((.........)))))))))).)))))).)))). ( -47.70) >DroSim_CAF1 11639 90 - 1 CGUGAUGGUGCGUCGAAGGUUGAUAGGGGUACCUAUAAGAACUUUUUCCUCUGUCAUAAC------------------------------UGGAGGUGCACCACGUGGUGUAUUCGAUGU .........(((((((((..(((((((((..................)))))))))...)------------------------------)....(((((((....)))))))))))))) ( -28.47) >DroEre_CAF1 11367 90 - 1 CGUAAUGGUGCGUCGAAGGUUGAUGGGGGUGCCUAUCAGCACUUUUUCCUCUGUCAUAAC------------------------------UGGAGGUGCACCACGUGGUGUAUUUGAUGG ..........(((((((.(((((((((....))))))))).......(((((((....))------------------------------.)))))((((((....))))))))))))). ( -31.10) >consensus CGUGAUGGUGCGUCGAAGGUUGAUGGGGGUACCUAUAAGAACUUUUUCCUCUGUCAUAAC______________________________UGGAGGUGCACCACGUGGUGUAUUCGAUGU ......(((((...(((((((.(((((....)))))...))))))).(((((.......................................))))).)))))((((.(......).)))) (-19.21 = -19.41 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:10 2006