| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
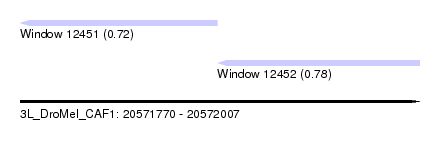
| Location | 20,571,770 – 20,572,007 |
| Length | 237 |
| Max. P | 0.781474 |

| Location | 20,571,770 – 20,571,887 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -48.20 |
| Consensus MFE | -31.38 |
| Energy contribution | -34.04 |
| Covariance contribution | 2.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20571770 117 - 23771897 CUCCUGGCAACCAGGAGGCCCCGGGGCUCCACCACCUCCACCAGGCGGGUUACCAGCGCCUCCCGGGGGAUU---GUUUAAUCGAUGUGGCCCGUUGCCGGGAUCAAUGUCAGUUGGUUU .((((((((((..((((.((....)))))).((((((((...(((((.........)))))...)))))(((---(......)))).)))...))))))))))(((((....)))))... ( -46.90) >DroSec_CAF1 16713 117 - 1 CUCCUGGCAACCAGGAGGCCCCGGGACUCCGCCACCUCCACCACCCGGGUUGCCAGCGCCUCCCGGGGGAUU---GUUUAAUCGAUGUGGCCCGUUGCCGGGAUCAAUGUCAGUUGGUUU .((((((((((..((((.((...)).))))(((((.....((.((((((..((....))..))))))))(((---(......)))))))))..))))))))))(((((....)))))... ( -53.50) >DroAna_CAF1 38291 99 - 1 CUCCUGGCAACCGGGGG---------------CAACU------CCUUGGUUUCCCUGGGCUCCCGGGGGAUUGUUGUUUAAUCGAUGUGUGCCGUUGCUGGGAUCAAAGUCUUUUGGCUU .(((..(((((.((((.---------------...))------))..(((..((((((....))))))(((((.....))))).......))))))))..)))...(((((....))))) ( -43.50) >DroSim_CAF1 10209 117 - 1 CUCCUGGCAACCAGGAGGCCCCGGGACUCCGCCACCUCCACCAGCCGGGUUGCCAGCGCCUCCCGGGGGAUU---GUUUAAUCGAUGUGGCCCGUUGCCGGGAUCAAUGUCAGUUGGUUU .((((((((((..((((.((...)).))))(((((.((..((..(((((..((....))..)))))))((((---....)))))).)))))..))))))))))(((((....)))))... ( -49.90) >DroEre_CAF1 9978 117 - 1 CUCCUGGCAACCAGGAGGCCCCUGGACUCCGCCACCUGCGCCACCCUGGUUGCCAGCACCAGCUUGGGGAUU---GUUUAAUCGAUGUGGCCCGUUGCCGGGAUCAAUGUCAGUUGGCUU ...((((((((((((.((((..(((......)))...).)))..))))))))))))..((((((....((((---....))))(((((((((((....)))).)).)))))))))))... ( -47.20) >consensus CUCCUGGCAACCAGGAGGCCCCGGGACUCCGCCACCUCCACCACCCGGGUUGCCAGCGCCUCCCGGGGGAUU___GUUUAAUCGAUGUGGCCCGUUGCCGGGAUCAAUGUCAGUUGGUUU .((((((((((..((((.((...)).))))(((((........((((((..((....))..)))))).((((.......))))...)))))..))))))))))(((((....)))))... (-31.38 = -34.04 + 2.66)

| Location | 20,571,887 – 20,572,007 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -44.76 |
| Consensus MFE | -37.24 |
| Energy contribution | -38.12 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20571887 120 - 23771897 CAGGACACUGGCACUUCCCAUGCCACUAAUGCUCUCGAUUUGACUCGCACCGUCGUUGAACCAAAUGUCCGGGGUAUGGGGAUUCUUGGGCGUAUCGCUCAGAUAGCCGGAUGUGUCCUC .(((((((((((..((((((((((.....(((..((.....))...)))(((.((((......))))..)))))))))))))...(((((((...)))))))...))))...))))))). ( -45.60) >DroSec_CAF1 16830 120 - 1 CAGGACACUGGCACUUCCCAUGCCACUAAUGCUCUCGAUUUGACUCGCACCGUCGUUGAACCAAAUGUCCGGAGUGUGGGGAUUCUUGGGCGUAUCGCUCAGAUAGCCGGAUGUGUCCUC .(((((((((((..(((((((((......(((..((.....))...)))(((.((((......))))..))).)))))))))...(((((((...)))))))...))))...))))))). ( -40.40) >DroAna_CAF1 38390 120 - 1 CAGGACACUGGCGCUUCCCAAGCCACUGACGCUCUCGAUUUGGCUCGCACCGUCGUUGAACCAAAUGUCGGGCGUGUGCGGAUUCUUGGGCGUGUCGCUAAGAUAGCCGGAAGUGUCCUC .(((((((((((((..((((((...(((((((.((((((((((.(((.((....))))).))))..)))))).)))).)))...)))))).)))))((((...))))....)))))))). ( -49.20) >DroSim_CAF1 10326 120 - 1 CAGGACACUGGCACUUCCCAUGCCACUAAUGCUCUCGAUUUGACUCGCACCGUCGUUGAACCAAAUGUCCGGGGUGUGGGGAUUCUUGGGCGUAUCGCUCAGAUAGCCGGAUGUGUCCUC .(((((((((((..((((((((((.....(((..((.....))...)))(((.((((......))))..)))))))))))))...(((((((...)))))))...))))...))))))). ( -44.60) >DroEre_CAF1 10095 120 - 1 CAGGACACUGGCACUACCCAUGCCACUAAUGCUCUCGAUUUGACUGGCCCCGUCGUUGAACCAAAUGUCCGGGGUAUGGGGAUUCUUGGGCGUAUCGCUCAGAUAGCCGGAUGUGUCCUC .(((((((((((....((((((((......(((.((.....))..))).(((.((((......))))..))))))))))).....(((((((...)))))))...))))...))))))). ( -44.00) >consensus CAGGACACUGGCACUUCCCAUGCCACUAAUGCUCUCGAUUUGACUCGCACCGUCGUUGAACCAAAUGUCCGGGGUGUGGGGAUUCUUGGGCGUAUCGCUCAGAUAGCCGGAUGUGUCCUC .(((((((((((..((((((((((..........(((((..(((.......)))))))).((........))))))))))))...(((((((...)))))))...))))...))))))). (-37.24 = -38.12 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:53:04 2006