
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,569,136 – 20,569,279 |
| Length | 143 |
| Max. P | 0.858317 |

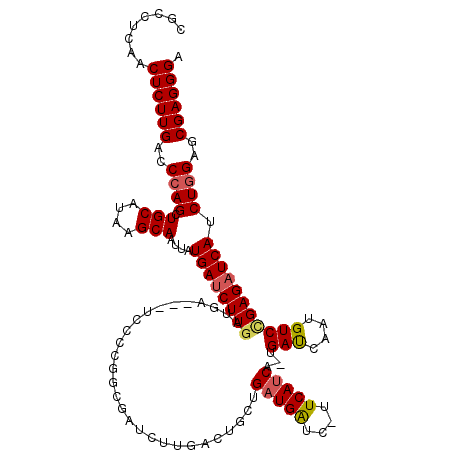
| Location | 20,569,136 – 20,569,251 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -34.64 |
| Consensus MFE | -25.60 |
| Energy contribution | -25.96 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20569136 115 + 23771897 CGCCUCAACUCUUGACCCAGCUGCAUAAGCAAUUAUGAUCUUUAUGA---UCCCCGGCGAUCUUGACUGUUGAUGAUC-UUCAUCA-UGAUCAAUGUCCGAGAUCAUCUAGAGCGAGGGA ..((((..((((.((....((((.((((....))))((((.....))---))..))))(((((((((.((((((.((.-......)-).)))))))).))))))).)).)))).)))).. ( -34.10) >DroSec_CAF1 13912 115 + 1 CGCCUCAACUCUUGACCCAGCUGCAUAAGCAAUUAUGAUCUUGAUGA---UCCCCGGCGAUCUUGACUGCUGAUGAUC-UUCAUCA-UGAUCAAUGUCUGAGAUCAUCUGGAGCGAGGGA ((((...............(((.....)))......((((.....))---))...)))).(((((.((.(.(((((((-((...((-(.....)))...))))))))).).))))))).. ( -34.50) >DroAna_CAF1 34937 108 + 1 CGCCACGACUCUUGACCCAGCUGCAUAAGCAAUUAUGAUCUUGAUGACCAUCCCC------------CCCAGAUGACCCAUCACCAGUGACCAAUGUCCGAGAUCAACUGGAGCGAGGGA ........((((((..((((.(((....)))....(((((((((((..((((...------------....))))...))))....(.(((....))))))))))).))))..)))))). ( -30.60) >DroSim_CAF1 7574 115 + 1 CGCCUCAACUCUUGACCCAGCUGCAUAAGCAAUUAUGAUCUUGAUGA---UCCCCGGCGAUCUUGACUGCUGAUGAUC-UUCAUCA-UGAUCAAUGUCCGAGAUCAUCUGGAGCGAGGGA ........((((((..((((.(((....)))...(((((((((....---....(((((........))))).(((((-.......-.))))).....)))))))))))))..)))))). ( -35.50) >DroEre_CAF1 7435 114 + 1 CGCCUCCACUCUUGACCCAGCUGCACAAGCAAUUAUGAGCUUGCUGA---UCC-CGGCUAGCUUGACUGCUGAUGGUC-UUCAUCA-UGAUCAAUGUCCGAGAUCAUCUGGAGCGAGGGA ........((((((..((((((((....))).....(((((.((((.---...-)))).)))))....)))(((((((-((...((-(.....)))...))))))))).))..)))))). ( -38.50) >consensus CGCCUCAACUCUUGACCCAGCUGCAUAAGCAAUUAUGAUCUUGAUGA___UCCCCGGCGAUCUUGACUGCUGAUGAUC_UUCAUCA_UGAUCAAUGUCCGAGAUCAUCUGGAGCGAGGGA ........((((((..((((.(((....)))....((((((((............................(((((....)))))...(((....))))))))))).))))..)))))). (-25.60 = -25.96 + 0.36)


| Location | 20,569,136 – 20,569,251 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -27.02 |
| Energy contribution | -29.86 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20569136 115 - 23771897 UCCCUCGCUCUAGAUGAUCUCGGACAUUGAUCA-UGAUGAA-GAUCAUCAACAGUCAAGAUCGCCGGGGA---UCAUAAAGAUCAUAAUUGCUUAUGCAGCUGGGUCAAGAGUUGAGGCG ..(((((((((..((((((((((...(((((..-(((((..-...)))))...))))).....)).))))---))))...((((....((((....))))...)))).)))).))))).. ( -39.20) >DroSec_CAF1 13912 115 - 1 UCCCUCGCUCCAGAUGAUCUCAGACAUUGAUCA-UGAUGAA-GAUCAUCAGCAGUCAAGAUCGCCGGGGA---UCAUCAAGAUCAUAAUUGCUUAUGCAGCUGGGUCAAGAGUUGAGGCG ..((((((((..(((((((((.((..(((((..-(((((..-...)))))...)))))..))....))))---)))))..((((....((((....))))...))))..))).))))).. ( -42.30) >DroAna_CAF1 34937 108 - 1 UCCCUCGCUCCAGUUGAUCUCGGACAUUGGUCACUGGUGAUGGGUCAUCUGGG------------GGGGAUGGUCAUCAAGAUCAUAAUUGCUUAUGCAGCUGGGUCAAGAGUCGUGGCG ((((((...((((.(((((.((..(((..(...)..))).))))))).)))))------------)))))(((((.....)))))....(((....)))((((.(.(....).).)))). ( -34.60) >DroSim_CAF1 7574 115 - 1 UCCCUCGCUCCAGAUGAUCUCGGACAUUGAUCA-UGAUGAA-GAUCAUCAGCAGUCAAGAUCGCCGGGGA---UCAUCAAGAUCAUAAUUGCUUAUGCAGCUGGGUCAAGAGUUGAGGCG ..((((((((..(((((((((((...(((((..-(((((..-...)))))...))))).....)).))))---)))))..((((....((((....))))...))))..))).))))).. ( -42.40) >DroEre_CAF1 7435 114 - 1 UCCCUCGCUCCAGAUGAUCUCGGACAUUGAUCA-UGAUGAA-GACCAUCAGCAGUCAAGCUAGCCG-GGA---UCAGCAAGCUCAUAAUUGCUUGUGCAGCUGGGUCAAGAGUGGAGGCG ((((((..(((((.(((((((((...(((((..-(((((..-...)))))...))))).....)))-)))---)))((((((........))))))....)))))....))).))).... ( -44.90) >consensus UCCCUCGCUCCAGAUGAUCUCGGACAUUGAUCA_UGAUGAA_GAUCAUCAGCAGUCAAGAUCGCCGGGGA___UCAUCAAGAUCAUAAUUGCUUAUGCAGCUGGGUCAAGAGUUGAGGCG ..((((((((..(((((((((((...(((((...(((((......)))))...))))).....))))))....)))))..((((....((((....))))...))))..)))).)))).. (-27.02 = -29.86 + 2.84)



| Location | 20,569,176 – 20,569,279 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -34.64 |
| Energy contribution | -34.83 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20569176 103 + 23771897 UUUAUGAUCCCCGGCGAUCUUGACUGUUGAUGAUCUUCAUCAUGAUCAAUGUCCGAGAUCAUCUAGAGCGAGGGAGCUGUGGCUGGGAUCUUGGGGGACUUCG .....((((((((((..(((((.((.(.(((((((((...(((.....)))...))))))))).).)))))))..)))).....))))))..((....))... ( -37.70) >DroSec_CAF1 13952 103 + 1 UUGAUGAUCCCCGGCGAUCUUGACUGCUGAUGAUCUUCAUCAUGAUCAAUGUCUGAGAUCAUCUGGAGCGAGGGAGCUGUGGCUGGGAUCUUGGGGGACUUCG .....((((((((((..(((((.((.(.(((((((((...(((.....)))...))))))))).).)))))))..)))).....))))))..((....))... ( -39.00) >DroSim_CAF1 7614 102 + 1 UUGAUGAUCCCCGGCGAUCUUGACUGCUGAUGAUCUUCAUCAUGAUCAAUGUCCGAGAUCAUCUGGAGCGAGGGAGCUGUGGCUGGGAUCU-GGGGGACUUCG .....((((((((((..(((((.((.(.(((((((((...(((.....)))...))))))))).).)))))))..)))).....)))))).-((....))... ( -39.00) >DroEre_CAF1 7475 102 + 1 UUGCUGAUCC-CGGCUAGCUUGACUGCUGAUGGUCUUCAUCAUGAUCAAUGUCCGAGAUCAUCUGGAGCGAGGGAGCUGCGGCUGGUAUCUUGGGGGACUUCG .....(((.(-((((((((((..((((((((((((((...(((.....)))...)))))))))...))).)).)))))..)))))).)))..((....))... ( -34.60) >consensus UUGAUGAUCCCCGGCGAUCUUGACUGCUGAUGAUCUUCAUCAUGAUCAAUGUCCGAGAUCAUCUGGAGCGAGGGAGCUGUGGCUGGGAUCUUGGGGGACUUCG .....((((((((((..(((((.((.(.(((((((((...(((.....)))...))))))))).).)))))))..)))).....))))))..((....))... (-34.64 = -34.83 + 0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:59 2006