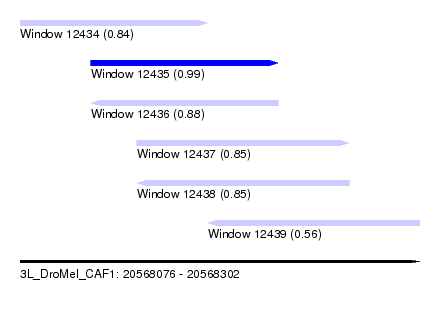
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,568,076 – 20,568,302 |
| Length | 226 |
| Max. P | 0.987783 |

| Location | 20,568,076 – 20,568,182 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20568076 106 + 23771897 CCCCGUCUCACAAAUUAGCUCCAUUACAACGCUGGAACGAUUGAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAAAAACGCAAAAUACAAA .................((((((.........))))..((((....))))..((((((..(((((((.....)))))))..)))))).......)).......... ( -24.50) >DroSec_CAF1 12856 106 + 1 CCCCGUCUCACAAAUUAGCUCCAUUACAUCGCACGAACGAUCCAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAGAAACGCAAAAUACAAA ..............................((.....(((....))).(((.((((((..(((((((.....)))))))..)))))).)))...)).......... ( -25.80) >DroSim_CAF1 6512 106 + 1 CCCCGUCUCACAAAUUAGCUCCAUUACAUCGCUCGAACGAUCCAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAGAAACGAAAAAUACAAA ............................(((.((((........))))(((.((((((..(((((((.....)))))))..)))))).)))..))).......... ( -25.70) >DroEre_CAF1 6420 106 + 1 CCCGGUCUCCCAAAUUAGCUCCAUUACAGAGUCCGUCCGAUCCAUCGAUCUUGCCUGUGAAAUCCAUUUCAAGUGGUUUCAGCAGCCCGAAAACGCAAAAUACAAG ..(((.(((...................))).)))...((((....))))((((((((((((.((((.....))))))))).))).........))))........ ( -19.41) >consensus CCCCGUCUCACAAAUUAGCUCCAUUACAUCGCUCGAACGAUCCAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAAAAACGCAAAAUACAAA .................((...................((((....))))..((((((..(((((((.....)))))))..)))))).......)).......... (-18.94 = -19.50 + 0.56)

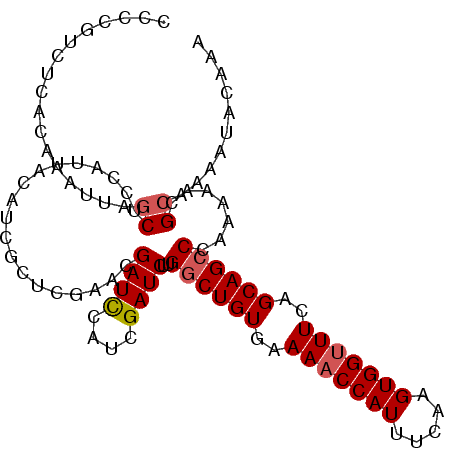
| Location | 20,568,116 – 20,568,222 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20568116 106 + 23771897 UUGAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAAAAACGCAAAAUACAAAAUACCCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACC ............((((((..(((((((.....)))))))..))))))......(((..(.(((((.......))))).)..)))....((((....))))...... ( -28.00) >DroSec_CAF1 12896 106 + 1 UCCAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAGAAACGCAAAAUACAAACCCCUCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACC ........(((.((((((..(((((((.....)))))))..)))))).)))..(((..(.(((((.......))))).)..)))....((((....))))...... ( -31.20) >DroSim_CAF1 6552 106 + 1 UCCAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAGAAACGAAAAAUACAAACCCCUCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACC ...((((.(((.((((((..(((((((.....)))))))..)))))).))).........(((((.......))))).....))))..((((....))))...... ( -29.90) >DroEre_CAF1 6460 104 + 1 UCCAUCGAUCUUGCCUGUGAAAUCCAUUUCAAGUGGUUUCAGCAGCCCGAAAACGCAAAAUACAAGCCC--CUUGUAUUGGGCGAUACUCUCACUUGAGAAUAACC ....(((...((((...(((((.((((.....)))))))))))))..)))...(((..((((((((...--))))))))..)))....((((....))))...... ( -27.10) >consensus UCCAUCGAUCUUGGCUGUGAAAACCAUUUCAAGUGGUUUCAGCAGCCCAAAAACGCAAAAUACAAACCCCUCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACC ...((((.((..((((((..(((((((.....)))))))..)))))).............(((((.......)))))..)).))))..((((....))))...... (-25.14 = -25.45 + 0.31)

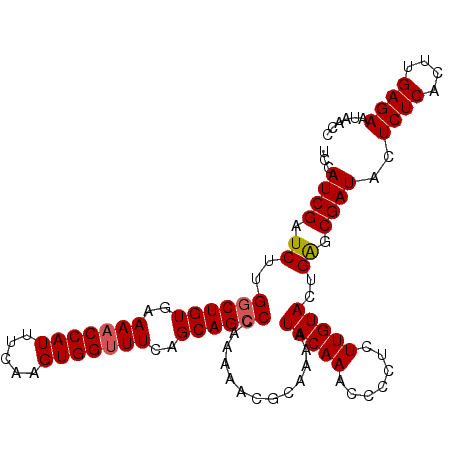
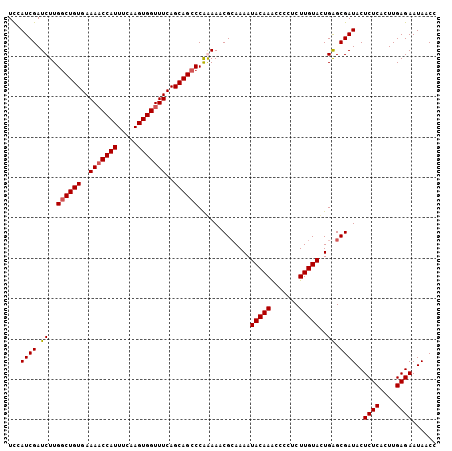
| Location | 20,568,116 – 20,568,222 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -28.45 |
| Energy contribution | -28.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20568116 106 - 23771897 GGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGGGUAUUUUGUAUUUUGCGUUUUUGGGCUGCUGAAACCACUUGAAAUGGUUUUCACAGCCAAGAUCGAUCAA ((((((((((....))))))..(((..((((((((.....))))))))..)))......(((((..(((((((.......)))))))..)))))..))))...... ( -31.60) >DroSec_CAF1 12896 106 - 1 GGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGGUUUGUAUUUUGCGUUUCUGGGCUGCUGAAACCACUUGAAAUGGUUUUCACAGCCAAGAUCGAUGGA ...(((((((....))))))).(((..((((((((.....))))))))..)))..(((.(((((..(((((((.......)))))))..))))).)))........ ( -33.70) >DroSim_CAF1 6552 106 - 1 GGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGGUUUGUAUUUUUCGUUUCUGGGCUGCUGAAACCACUUGAAAUGGUUUUCACAGCCAAGAUCGAUGGA (((.((((((....)))))))))....((((((((.....))))))))..(((..(((.(((((..(((((((.......)))))))..))))).))).))).... ( -31.60) >DroEre_CAF1 6460 104 - 1 GGUUAUUCUCAAGUGAGAGUAUCGCCCAAUACAAG--GGGCUUGUAUUUUGCGUUUUCGGGCUGCUGAAACCACUUGAAAUGGAUUUCACAGGCAAGAUCGAUGGA (((.((((((....)))))))))((((........--))))(((.((((((((.((((((....)))))).).(((((((....))))).)))))))))))).... ( -28.90) >consensus GGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGGUUUGUAUUUUGCGUUUCUGGGCUGCUGAAACCACUUGAAAUGGUUUUCACAGCCAAGAUCGAUGGA ...(((((((....))))))).(((..((((((((.....))))))))..)))..(((.(((((.((((((((.......))).)))))))))).)))........ (-28.45 = -28.57 + 0.12)

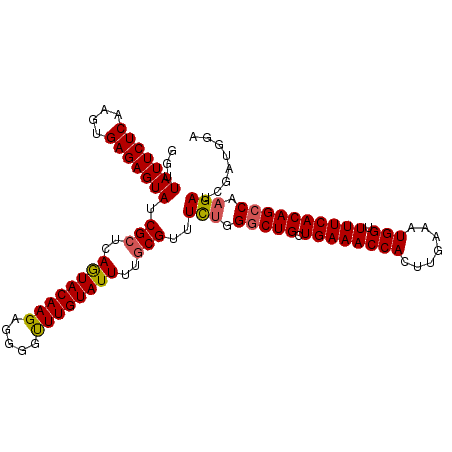
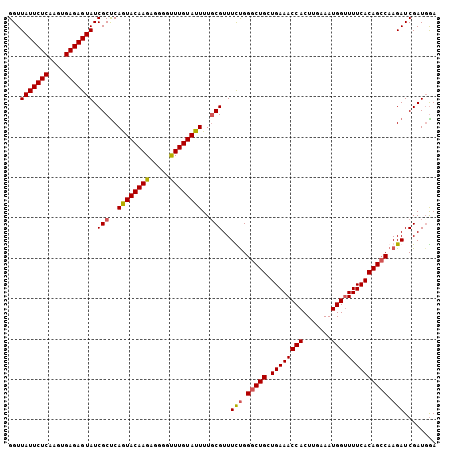
| Location | 20,568,142 – 20,568,262 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -24.83 |
| Energy contribution | -25.20 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.850058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20568142 120 + 23771897 UUUCAAGUGGUUUCAGCAGCCCAAAAACGCAAAAUACAAAAUACCCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACCCUAGAGUCGUCGUUUGCUACACGAGUUGCCCAAAUCACUU ....(((((((((..(((((.......(((..(.(((((.......))))).)..)))....((((....))))............((((.((.....)))))))))))..))))))))) ( -27.00) >DroSec_CAF1 12922 120 + 1 UUUCAAGUGGUUUCAGCAGCCCAGAAACGCAAAAUACAAACCCCUCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACCCUAGAGUCGUCGUUUGCUACACGAGUUGCCCAAAUCACUU ....(((((((((..(((((........)).............(((.((((..((((((((((((.................))))).)))))))..)))).))).)))..))))))))) ( -29.43) >DroSim_CAF1 6578 120 + 1 UUUCAAGUGGUUUCAGCAGCCCAGAAACGAAAAAUACAAACCCCUCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACCCUAGAGUCGUCGUUUGCUACACGAGUUGCCCGAAUCACUU ....(((((((((..(((((.(......)...............((.((((..((((((((((((.................))))).)))))))..)))).)))))))..))))))))) ( -27.73) >DroEre_CAF1 6486 115 + 1 UUUCAAGUGGUUUCAGCAGCCCGAAAACGCAAAAUACAAGCCC--CUUGUAUUGGGCGAUACUCUCACUUGAGAAUAACCCUAGAGUC---GUUUGCUACACGAGUUGCCCAAAUCACUU ....(((((((((..(((((.((.....((((((((((((...--))))))))(((..((.(((......))).))..))).......---..))))....)).)))))..))))))))) ( -33.50) >consensus UUUCAAGUGGUUUCAGCAGCCCAAAAACGCAAAAUACAAACCCCUCUUGUACUGAGCGAUACUCUCACUUGAGAAUAACCCUAGAGUCGUCGUUUGCUACACGAGUUGCCCAAAUCACUU ....(((((((((..(((((.(............(((((.......)))))..((((((((((((.................))))).))))))).......).)))))..))))))))) (-24.83 = -25.20 + 0.38)

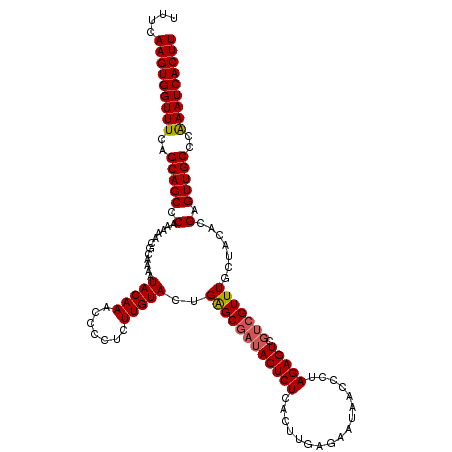
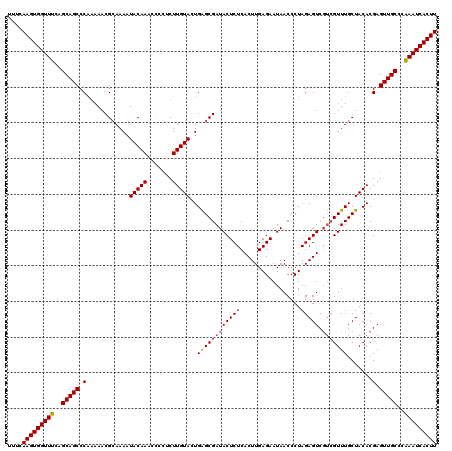
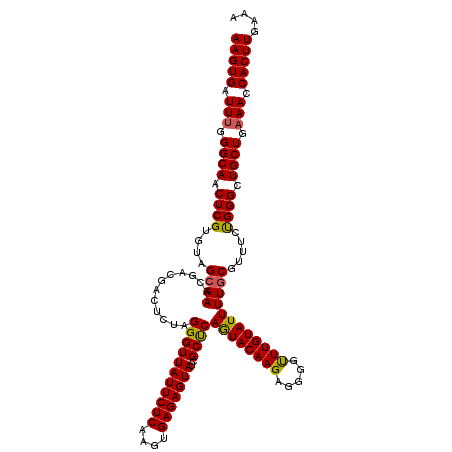
| Location | 20,568,142 – 20,568,262 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -34.08 |
| Energy contribution | -33.83 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20568142 120 - 23771897 AAGUGAUUUGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGGGUAUUUUGUAUUUUGCGUUUUUGGGCUGCUGAAACCACUUGAAA (((((.(((.((((.((((....(((((..((((.....(((((((((((....)))))))..))))(((((....)))))))))..))))).....)))).)))).))).))))).... ( -35.50) >DroSec_CAF1 12922 120 - 1 AAGUGAUUUGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGGUUUGUAUUUUGCGUUUCUGGGCUGCUGAAACCACUUGAAA (((((.(((.((((....((.(((.(((((.(((.....(((((((((((....)))))))..))))((((((((.....))))))))))))))))))).)))))).))).))))).... ( -35.30) >DroSim_CAF1 6578 120 - 1 AAGUGAUUCGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGGUUUGUAUUUUUCGUUUCUGGGCUGCUGAAACCACUUGAAA (((((.((..((((....((.(((.((((((........(((((((((((....)))))))..))))((((((((.....))))))))..))))))))).))))))..)).))))).... ( -32.40) >DroEre_CAF1 6486 115 - 1 AAGUGAUUUGGGCAACUCGUGUAGCAAAC---GACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCCCAAUACAAG--GGGCUUGUAUUUUGCGUUUUCGGGCUGCUGAAACCACUUGAAA (((((.(((.((((.((((....((((..---.......(((((((((((....)))))))..))))((((((((--...)))))))))))).....)))).)))).))).))))).... ( -40.70) >consensus AAGUGAUUUGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGGUUUGUAUUUUGCGUUUCUGGGCUGCUGAAACCACUUGAAA (((((.(((.((((.((((....((((............(((((((((((....)))))))..))))((((((((.....)))))))))))).....)))).)))).))).))))).... (-34.08 = -33.83 + -0.25)

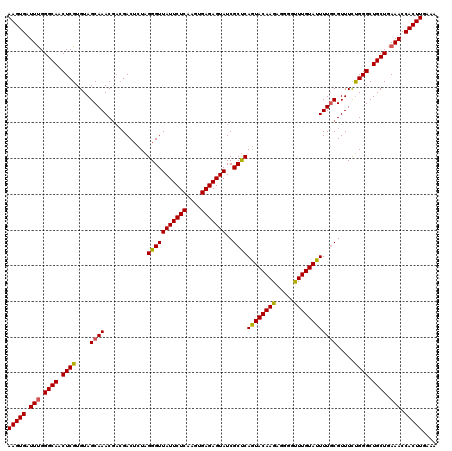
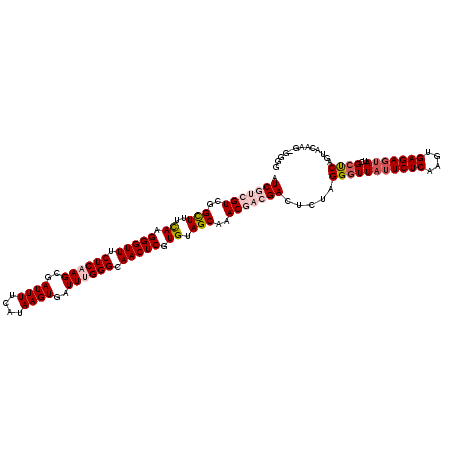
| Location | 20,568,182 – 20,568,302 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.54 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -28.46 |
| Energy contribution | -30.18 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20568182 120 - 23771897 AUCGUCGUCGGCUUUCAAGGGUUUCUCAAGCGAUUUUCAUAAGUGAUUUGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGGGUAU .(((((((..(((..((.(((((.((((((..((((....))))..)))))).))))).)).)))..))))))).....(((((((((((....)))))))..)))).((((....)))) ( -38.30) >DroSec_CAF1 12962 120 - 1 AUCGUCGUCGGUUUUUAAGGGUUUCUCAAGCGAUUUUCAUAAGUGAUUUGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGG .(((((((..(((.....(((((.((((((..((((....))))..)))))).)))))....)))..)))))))((((((.(.(((((((....))))))).).)).......))))... ( -36.01) >DroAna_CAF1 34083 107 - 1 AUCGUCGUCUGCUUACAAGGGUUUCUCAAGCGAUUUUCAUAAGUGAUUCGGGCAACUCGUGUAGCAAAC---GACUUAAGGUUUCAUCUCAAGUGAGAGUAUAGUUCAAU---------- ...(((((.((((.(((.(((((.((....(((...(((....))).))))).))))).))))))).))---)))...........((((....))))............---------- ( -26.20) >DroSim_CAF1 6618 120 - 1 AUCGUCGUCGGCUUUCAAGGGUUUCUCAAGCGAUUUUCAUAAGUGAUUCGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAGAGGGG .(((((((..(((..((.(((((.((....(((...(((....))).))))).))))).)).)))..)))))))((((((.(.(((((((....))))))).).)).......))))... ( -35.41) >DroEre_CAF1 6526 115 - 1 AUCGUCGUCGGCUUUCAAGGGUUUCUCAAGCGAUUUUUAUAAGUGAUUUGGGCAACUCGUGUAGCAAAC---GACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCCCAAUACAAG--GGG ...(((((..(((..((.(((((.((((((..((((....))))..)))))).))))).)).)))..))---)))....(((((((((((....)))))))..))))........--... ( -37.20) >consensus AUCGUCGUCGGCUUUCAAGGGUUUCUCAAGCGAUUUUCAUAAGUGAUUUGGGCAACUCGUGUAGCAAACGACGACUCUAGGGUUAUUCUCAAGUGAGAGUAUCGCUCAGUACAAG_GGGG .(((((((..(((..((.(((((.((((((..((((....))))..)))))).))))).)).)))..))))))).....(((((((((((....)))))))..))))............. (-28.46 = -30.18 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:51 2006