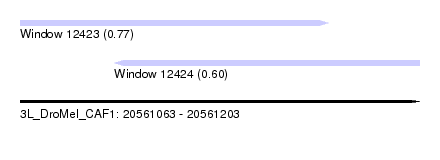
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,561,063 – 20,561,203 |
| Length | 140 |
| Max. P | 0.771557 |

| Location | 20,561,063 – 20,561,171 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 96.97 |
| Mean single sequence MFE | -40.63 |
| Consensus MFE | -34.87 |
| Energy contribution | -34.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20561063 108 + 23771897 ACUCGGGCCAAGACAAAGGCAAUGUCGGCCAGGAAAGUGGCAAGUCAAAGGCAGUGGCAGUGGCAUCUGCAUUAUGGCAAUGCGGAGUAUUGUGUUGCCUC---UUUCCCA .....((((..((((.......)))))))).((((((.(((((((((...((....))..)))).((((((((.....))))))))........))))).)---))))).. ( -41.60) >DroSim_CAF1 104446 111 + 1 ACUCCGGCCAAGACAAAGGCAAUGUCGGCCAGGAAAGUGGCAAGUCAAAGGCAGUGGCAGUCGCAUCUGCAUUAUGGCAAUGCGGAGUAUUGUGUUGCCUCUUCUUUCCCA .....((((..((((.......)))))))).((((((.(((((((.....))....(((((.((.((((((((.....))))))))))))))).)))))....)))))).. ( -38.40) >DroEre_CAF1 105618 108 + 1 ACUCCGGCCAAGACAAAGGCAAUGUCGGCCAGGAAAGUGGCAAGUCAAAGGCAGUGGCAGUGGCAUCUGCAUUAUGGCAAUGCGGAGUAUUGUGUUGCCUC---UUUCCCA .....((((..((((.......)))))))).((((((.(((((((((...((....))..)))).((((((((.....))))))))........))))).)---))))).. ( -41.90) >consensus ACUCCGGCCAAGACAAAGGCAAUGUCGGCCAGGAAAGUGGCAAGUCAAAGGCAGUGGCAGUGGCAUCUGCAUUAUGGCAAUGCGGAGUAUUGUGUUGCCUC___UUUCCCA .....((((..((((.......)))))))).(((((..(((((((.....))....(((((.((.((((((((.....))))))))))))))).))))).....))))).. (-34.87 = -34.87 + 0.00)



| Location | 20,561,096 – 20,561,203 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -29.91 |
| Energy contribution | -30.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20561096 107 - 23771897 -------GC-GGCAACAGGCAACCUGGCUAACUGGCAACAUGGGAAA---GAGGCAACACAAUACUCCGCAUUGCCAUAAUGCAGAUGCCACUGCCACUGCCUUUGACUUGCCACUUU -------..-(((((.(((((...((((....(((((.(...(((..---(.(....).).....)))((((((...)))))).).)))))..)))).))))).....)))))..... ( -32.20) >DroSim_CAF1 104479 110 - 1 -------GC-GGCAACAGGCAACCUGGCUAACUGGCAACAUGGGAAAGAAGAGGCAACACAAUACUCCGCAUUGCCAUAAUGCAGAUGCGACUGCCACUGCCUUUGACUUGCCACUUU -------..-(((((.(((((...((((....(((((..((((((.......(....).......))).))))))))...(((....)))...)))).))))).....)))))..... ( -31.54) >DroEre_CAF1 105651 115 - 1 CUGGCAACCUGGCAACUGGCAACCUGGCUAACUGGCAACAUGGGAAA---GAGGCAACACAAUACUCCGCAUUGCCAUAAUGCAGAUGCCACUGCCACUGCCUUUGACUUGCCACUUU .((((((...((((..(((((...((((...(((.((..((((.((.---(((...........)))....)).))))..)))))..)))).))))).))))......)))))).... ( -34.90) >consensus _______GC_GGCAACAGGCAACCUGGCUAACUGGCAACAUGGGAAA___GAGGCAACACAAUACUCCGCAUUGCCAUAAUGCAGAUGCCACUGCCACUGCCUUUGACUUGCCACUUU ..........(((((.(((((...((((....(((((.(...(((.......(....).......)))((((((...)))))).).)))))..)))).))))).....)))))..... (-29.91 = -30.57 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:37 2006