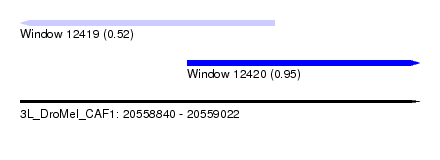
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,558,840 – 20,559,022 |
| Length | 182 |
| Max. P | 0.952597 |

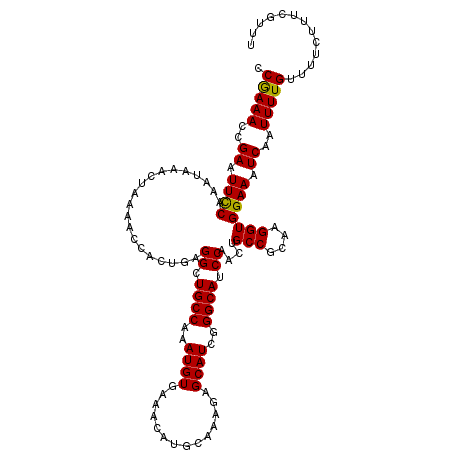
| Location | 20,558,840 – 20,558,956 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -25.15 |
| Energy contribution | -24.71 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20558840 116 - 23771897 CCGAAACCGAAUUCCAAAUAAACUAAAACCACUGAGGCUGCCAAAUGUGAAACAUGCAAAGAGCAUCGGGCAUCCAACUGCCGCAAGGUGGAAAUCAAUUUUGGUUUCUUUCGUUU ..(((((((((........................((.((((..((((..............))))..)))).))..(..((....))..)........)))))))))........ ( -31.34) >DroSim_CAF1 102204 116 - 1 CCGAAACCGAAUUCCAAAUAAACCAAAACCACUGAGGCUGCCAAAUGUGAAACAUGCAAAGAGCAUCGGGCAUCCAACUGCCGCAAGGUGGAAAUCAAUUUUGUUUUCUUUCGUUU .(((((..(((..(.((((................((.((((..((((..............))))..)))).))..(..((....))..)......)))).)..))))))))... ( -26.04) >DroEre_CAF1 103419 116 - 1 CCAAAACCGAAUUUCAAAUAAACUAAGUCCACUCAGGCUGCCAAAUGUAAAACAUGCAAAGAGCAUCGGGCAUCCAAGUGCCGCAAGGUGGAAAUCAAUUUUGUUUUCUUUCGUUU .......((((....(((((((.....(((.....((.((((..((((..............))))..)))).))....(((....)))))).......)))))))...))))... ( -25.44) >consensus CCGAAACCGAAUUCCAAAUAAACUAAAACCACUGAGGCUGCCAAAUGUGAAACAUGCAAAGAGCAUCGGGCAUCCAACUGCCGCAAGGUGGAAAUCAAUUUUGUUUUCUUUCGUUU .(((((..((.((((....................((.((((..((((..............))))..)))).))....(((....))))))).))..)))))............. (-25.15 = -24.71 + -0.44)


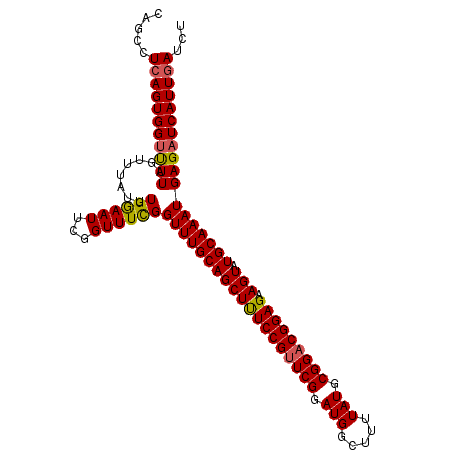
| Location | 20,558,916 – 20,559,022 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -32.31 |
| Energy contribution | -32.43 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20558916 106 + 23771897 CAGCCUCAGUGGUUUUAGUUUAUUUGGAAUUCGGUUUCGGUUUGCAGCUUUCCGUUCGGAUGGCUUUUAUGCGGACGGAGAAGUAUGCAAAUGAGAUCAUUGAUCU .....(((((((((((........((((((...))))))(((((((((((((((((((.(((.....))).))))))))).))).))))))))))))))))))... ( -35.40) >DroSim_CAF1 102280 106 + 1 CAGCCUCAGUGGUUUUGGUUUAUUUGGAAUUCGGUUUCGGUUUGCAGCUUUCCGUUCGGAUGGCUUUUAUGCGGACGGAGAAGUAUGCAAAUGAGAUCAUUGAUCU .....(((((((((((........((((((...))))))(((((((((((((((((((.(((.....))).))))))))).))).))))))))))))))))))... ( -35.40) >DroEre_CAF1 103495 106 + 1 CAGCCUGAGUGGACUUAGUUUAUUUGAAAUUCGGUUUUGGUUUGCAGCUCUCCGUUCGGAUGGCUUUUAUGCGGUCGGAGAAGUAUGCAAAUGAGAUCAUUGAUCU ....(((((....)))))............((((((((.(((((((((((((((..((.(((.....))).))..))))).))).))))))).)))..)))))... ( -29.60) >consensus CAGCCUCAGUGGUUUUAGUUUAUUUGGAAUUCGGUUUCGGUUUGCAGCUUUCCGUUCGGAUGGCUUUUAUGCGGACGGAGAAGUAUGCAAAUGAGAUCAUUGAUCU .....(((((((((((........((((((...))))))(((((((((((((((((((.(((.....))).))))))))).))).))))))))))))))))))... (-32.31 = -32.43 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:33 2006