| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,555,677 – 20,555,790 |
| Length | 113 |
| Max. P | 0.609238 |

| Location | 20,555,677 – 20,555,790 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.34 |
| Mean single sequence MFE | -35.61 |
| Consensus MFE | -15.37 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
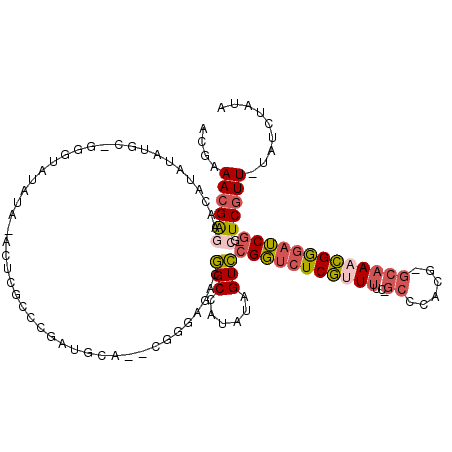
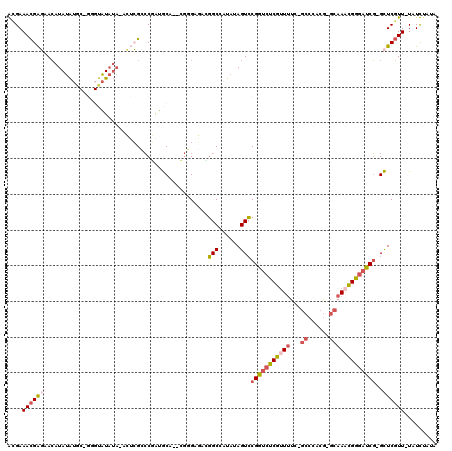
>3L_DroMel_CAF1 20555677 113 - 23771897 GCGAAACGAGAACAUAUAUGCCGGGUAUAUA-ACUCGCCCGAUGCA--CGGGAGAUGGCCUUAUAGUCCGGUCUCGUUUUC-GCCCACG-GCAAACGGGAUCG-GCUCGUU-UAUCUAAA ...(((((((....((((.(((((((.....-)))).((((.....--))))....)))..))))..((((((((((((..-(((...)-)))))))))))))-)))))))-)....... ( -41.90) >DroVir_CAF1 105854 102 - 1 AUGAAAUGAGUAAAUAAGUUC-GCGUUU----AGUCGAGCAAAGC------GG---CGCUACAUAGUGCGCACUCCCUCUU-ACCCACA--CAAUGGAUAGCU-GCUCUUUAUAUUUAUA .........(((((((.((((-((....----.).)))))..(((------((---(....(((.(((.(...........-...).))--).)))....)))-))).....))))))). ( -22.24) >DroPse_CAF1 116818 111 - 1 AUGAAACGGUAACAUAUAU-------AUAUACUGGGGGCCUGUACAUAAGCGACAAGGCAAUAUAGUCCGGUCUCGUUUUG-GCCCACGGGCAAACGGGAUCGCGUUCGUU-UAUUUAUA .......(((..(.(((..-------..)))...)..)))((((.(((((((((..(((......)))((((((((((...-(((....))).)))))))))).).)))))-))).)))) ( -32.10) >DroSim_CAF1 99019 113 - 1 GCGAAACGAGAACAUAUAUGCCGGGUAUAUA-ACUCGCCCGAUGCA--CGGGAGACGGCCUUAUAGUCCGGUCUCGUUUUC-GCCCACG-GCAAACGGGAUCG-GCUCGUU-UAUCUAUA ...(((((((....((((.(((((((.....-)))).((((.....--))))....)))..))))..((((((((((((..-(((...)-)))))))))))))-)))))))-)....... ( -42.20) >DroEre_CAF1 100120 114 - 1 GCGAAACGAGAACAUAUAUGCCGGGUAUAUAAACUCGCCCGAUGCA--CGGGAGACGGCCAUAUAGUCCGGUCUCGUUUUC-GCCCACG-GCAAACGGGAUCG-GCUCGUU-UAUCUAUA ...(((((((....((((((((((((......)))).((((.....--))))....)).))))))..((((((((((((..-(((...)-)))))))))))))-)))))))-)....... ( -43.90) >DroPer_CAF1 118807 114 - 1 AUGAAACGGUAACAUAUAUA-----UAUAUACAGGGGGCCUGUACAUAAGCGACAUGGCAAUAUAGUCCGGUCUCGUUUUGGGCCCACGGGCAACCGGGAUCGCGUUCGUU-UAUUUAUA ...((((((..((.......-----(((.((((((...)))))).))).((((............((((((.......))))))((.(((....))))).)))))))))))-)....... ( -31.30) >consensus ACGAAACGAGAACAUAUAUGC_GGGUAUAUA_ACUCGCCCGAUGCA__CGGGAGACGGCCAUAUAGUCCGGUCUCGUUUUC_GCCCACG_GCAAACGGGAUCG_GCUCGUU_UAUCUAUA ....((((((..............................................(((......)))(((((((((((...((......)))))))))))))..))))))......... (-15.37 = -16.02 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:30 2006