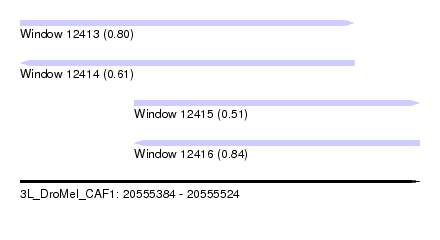
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,555,384 – 20,555,524 |
| Length | 140 |
| Max. P | 0.840063 |

| Location | 20,555,384 – 20,555,501 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -22.71 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20555384 117 + 23771897 ACAUCAUUAUUUUUCCAGUUGCUCACUUUGAAUAUUGCACUCGAUUGUUUUUCUCAAUCGCUCACCAUUUUGUGCGGCUUUUGAGUGCUUCACUCACAGGGCACACACUCUCGUGAA ...................(((((....(((.....(((((((((((.......)))))((((((......))).)))....)))))).....)))..)))))(((......))).. ( -27.40) >DroSim_CAF1 98726 117 + 1 ACAUCACUAUUUUUCCAGUUGCUCACUUUGAAUAUUGCACUCGAUUGUUUUUCUCGAUCUCUCACCAUUUUGUGCGGCAUUUGAGUGCUUCACUCACAGGGCACACACACUCGUGAA ...................(((((....(((.....(((((((((((.......)))))((.(((......))).)).....)))))).....)))..)))))(((......))).. ( -23.50) >DroEre_CAF1 99828 116 + 1 A-AACAAUAUUUUUCCAGUUGCUCACUUUGAAUAUUGCACUCGAUCGUUUUUCACGAUCUCUCACCAUUUUGUGCGGCAUUUGAGUGCUUCACUCACUGGUCACACACACACGUGAA .-..((((((((....(((.....)))..)))))))).....((((((.....))))))..((((.....((((.(((...((((((...))))))...))).)))).....)))). ( -29.50) >consensus ACAUCAAUAUUUUUCCAGUUGCUCACUUUGAAUAUUGCACUCGAUUGUUUUUCUCGAUCUCUCACCAUUUUGUGCGGCAUUUGAGUGCUUCACUCACAGGGCACACACACUCGUGAA ...................(((((....(((.....(((((((((((.......))))).(.(((......))).)......)))))).....)))..)))))(((......))).. (-22.71 = -22.60 + -0.11)

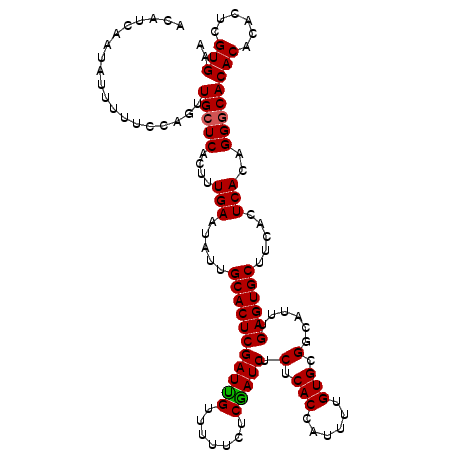
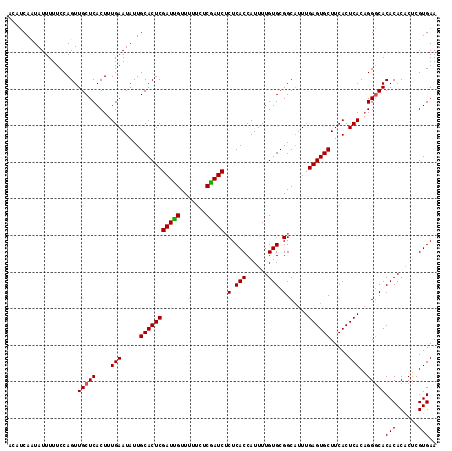
| Location | 20,555,384 – 20,555,501 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -31.16 |
| Consensus MFE | -25.05 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20555384 117 - 23771897 UUCACGAGAGUGUGUGCCCUGUGAGUGAAGCACUCAAAAGCCGCACAAAAUGGUGAGCGAUUGAGAAAAACAAUCGAGUGCAAUAUUCAAAGUGAGCAACUGGAAAAAUAAUGAUGU ..(((....)))..((((((.((((((..((((((....(((((........))).))(((((.......)))))))))))..)))))).)).).)))................... ( -32.30) >DroSim_CAF1 98726 117 - 1 UUCACGAGUGUGUGUGCCCUGUGAGUGAAGCACUCAAAUGCCGCACAAAAUGGUGAGAGAUCGAGAAAAACAAUCGAGUGCAAUAUUCAAAGUGAGCAACUGGAAAAAUAGUGAUGU (((((.(.((((((.((....((((((...))))))...))))))))...).)))))..((((..........((.(((((..............)).))).)).......)))).. ( -27.47) >DroEre_CAF1 99828 116 - 1 UUCACGUGUGUGUGUGACCAGUGAGUGAAGCACUCAAAUGCCGCACAAAAUGGUGAGAGAUCGUGAAAAACGAUCGAGUGCAAUAUUCAAAGUGAGCAACUGGAAAAAUAUUGUU-U .(((((......)))))((((((((((..((((((....((((.......))))....((((((.....))))))))))))..)))))...(....).)))))............-. ( -33.70) >consensus UUCACGAGUGUGUGUGCCCUGUGAGUGAAGCACUCAAAUGCCGCACAAAAUGGUGAGAGAUCGAGAAAAACAAUCGAGUGCAAUAUUCAAAGUGAGCAACUGGAAAAAUAAUGAUGU ..(((....)))(((......((((((..((((((....((((.......))))....(((((.......)))))))))))..))))))......)))................... (-25.05 = -25.17 + 0.11)

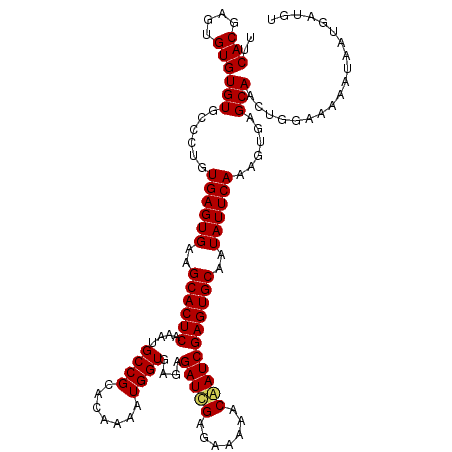
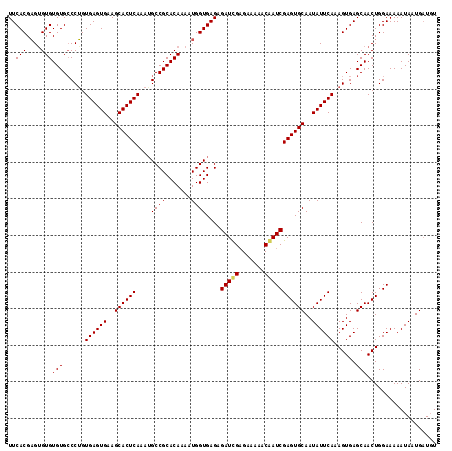
| Location | 20,555,424 – 20,555,524 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -20.46 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20555424 100 + 23771897 UCGAUUGUUUUUCUCAAUCGCUCACCAUUUUGUGCGGCUUUUGAGUGCUUCACUCACAGGGCACACACUCUCGUGAAUAUAUCUACUCGAAGAAUUCGUU .((((((.......)))))).((((.....((((.(.(((.((((((...)))))).))).).)))).....)))).....(((......)))....... ( -24.20) >DroSim_CAF1 98766 100 + 1 UCGAUUGUUUUUCUCGAUCUCUCACCAUUUUGUGCGGCAUUUGAGUGCUUCACUCACAGGGCACACACACUCGUGAAUAUAUCUACUCGAAGAAUUCGUU .(((..(((((((..(((...((((.....((((.(.(...((((((...))))))...).).)))).....))))....))).....)))))))))).. ( -20.90) >DroEre_CAF1 99867 100 + 1 UCGAUCGUUUUUCACGAUCUCUCACCAUUUUGUGCGGCAUUUGAGUGCUUCACUCACUGGUCACACACACACGUGAAUACAUCUACUCGAGGAAUUCGUU ..((((((.....))))))..((((.....((((.(((...((((((...))))))...))).)))).....)))).....(((......)))....... ( -25.00) >consensus UCGAUUGUUUUUCUCGAUCUCUCACCAUUUUGUGCGGCAUUUGAGUGCUUCACUCACAGGGCACACACACUCGUGAAUAUAUCUACUCGAAGAAUUCGUU ..(((((.......)))))..((((.....((((.(.(...((((((...))))))...).).)))).....)))).....(((......)))....... (-20.46 = -19.80 + -0.66)

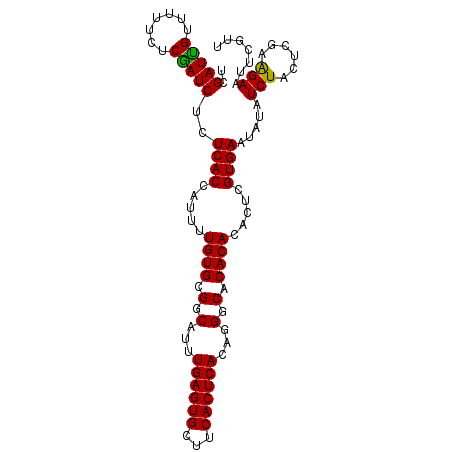
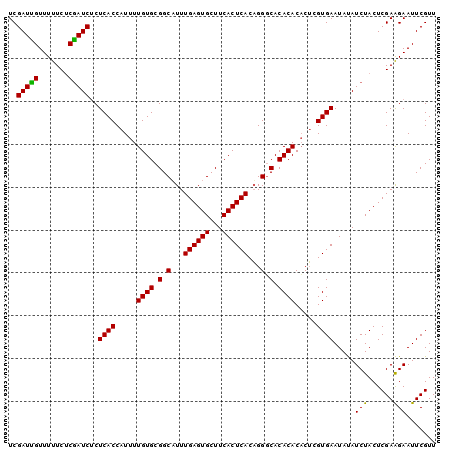
| Location | 20,555,424 – 20,555,524 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -24.55 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20555424 100 - 23771897 AACGAAUUCUUCGAGUAGAUAUAUUCACGAGAGUGUGUGCCCUGUGAGUGAAGCACUCAAAAGCCGCACAAAAUGGUGAGCGAUUGAGAAAAACAAUCGA ..(((.....)))..........(((((.(...((((((.(...((((((...))))))...).))))))...).)))))((((((.......)))))). ( -27.40) >DroSim_CAF1 98766 100 - 1 AACGAAUUCUUCGAGUAGAUAUAUUCACGAGUGUGUGUGCCCUGUGAGUGAAGCACUCAAAUGCCGCACAAAAUGGUGAGAGAUCGAGAAAAACAAUCGA ..(((.(((((.(((((....)))))((.(.((((((.((....((((((...))))))...))))))))...).))))))).))).............. ( -27.40) >DroEre_CAF1 99867 100 - 1 AACGAAUUCCUCGAGUAGAUGUAUUCACGUGUGUGUGUGACCAGUGAGUGAAGCACUCAAAUGCCGCACAAAAUGGUGAGAGAUCGUGAAAAACGAUCGA .........((((((((....))))).(((...((((((.(...((((((...))))))...).))))))..)))..))).((((((.....)))))).. ( -30.10) >consensus AACGAAUUCUUCGAGUAGAUAUAUUCACGAGUGUGUGUGCCCUGUGAGUGAAGCACUCAAAUGCCGCACAAAAUGGUGAGAGAUCGAGAAAAACAAUCGA ..(((.....)))..........(((((.(...((((((.(...((((((...))))))...).))))))...).))))).(((((.......))))).. (-24.55 = -25.00 + 0.45)

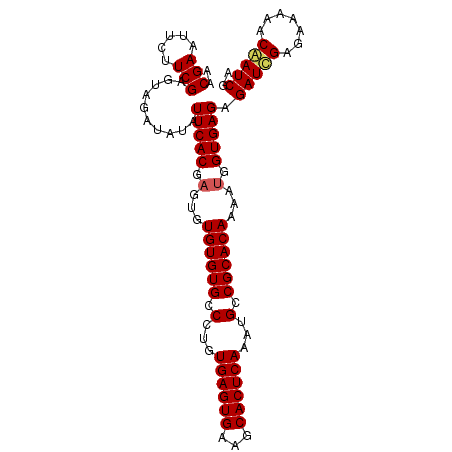
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:29 2006