
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,554,006 – 20,554,134 |
| Length | 128 |
| Max. P | 0.995672 |

| Location | 20,554,006 – 20,554,098 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -19.18 |
| Energy contribution | -18.96 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20554006 92 - 23771897 GCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCCAUAAAAACUCGCAUUCAAAACACAGAAGGCGUAAAAACAGCACCAAAUAAAGU ((.(((.((((((.((.(((......))).)).)))))).............(((........))))))((....)).))............ ( -24.70) >DroSec_CAF1 95868 92 - 1 GCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCUAUAAAAAAUCGCAUUAAAAACACAGAAAGCGUAAAAAUAGCACCAAAUAAAGU ((.....((((((.((.(((......))).)).)))))).........(((................)))........))............ ( -18.29) >DroEre_CAF1 98438 91 - 1 GCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCUAUAAA-AAUCGCAUCAAAAAUACAGAAAGCGUAAAAACAGCUCAAAAUAAAGU (.(((..((((((.((.(((......))).)).)))))).....-...(((................)))........)))).......... ( -18.09) >consensus GCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCUAUAAAAAAUCGCAUUAAAAACACAGAAAGCGUAAAAACAGCACCAAAUAAAGU ((.....((((((.((.(((......))).)).)))))).........(((................)))........))............ (-19.18 = -18.96 + -0.22)



| Location | 20,554,042 – 20,554,134 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20554042 92 + 23771897 UGAAUGCGAGUUUUUAUGGAGUGACCACACCUUUGUGUGAGGGCACUCCUGGCUGCGCACGCGCACUUGCCGUAAAUUUCUUUAUUUCCGCU .....(((.........((((((.((((((....))))).)..)))))).(((((((....))))...))).................))). ( -29.40) >DroSec_CAF1 95904 92 + 1 UUAAUGCGAUUUUUUAUAGAGUGACCACACCUUUGUGUGAGGGCACUCCUGGCUGCGCACGCGCACUUGCCGUAAAUUUCUUUCUUUCCGCU .....(((..........(((((.((((((....))))).)..)))))..(((((((....))))...))).................))). ( -25.90) >DroSim_CAF1 97395 92 + 1 UUAAUGCGAUUUUUUAUAGAGGGACCACACCUUUGUGUGAGGGCACUCCUGGCUGCGCACGCGCACUUGCCGUAAAUUUCUUUAUUUCCGCU .....(((.......((((((..(.(((((....)))))..((((.....((.((((....)))))))))).....)..))))))...))). ( -24.00) >DroEre_CAF1 98474 90 + 1 UUGAUGCGAUU-UUUAUAGAGUGACCACACCUUUGUGUGAGGGCACUCCUGGCUGCGCACGCGCACUUGCCGCAAAUUUCU-UAUUUCCGCU .....(((...-......(((((.((((((....))))).)..)))))..(((((((....))))...)))..........-......))). ( -25.90) >consensus UUAAUGCGAUUUUUUAUAGAGUGACCACACCUUUGUGUGAGGGCACUCCUGGCUGCGCACGCGCACUUGCCGUAAAUUUCUUUAUUUCCGCU .....(((..........(((((.((((((....))))).)..)))))..(((((((....))))...))).................))). (-24.52 = -24.78 + 0.25)

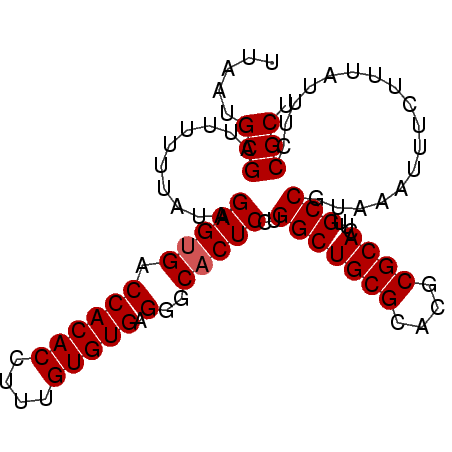
| Location | 20,554,042 – 20,554,134 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20554042 92 - 23771897 AGCGGAAAUAAAGAAAUUUACGGCAAGUGCGCGUGCGCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCCAUAAAAACUCGCAUUCA .((((...((((....)))).(((...((((....))))))).((((((.((.(((......))).)).))))))........))))..... ( -32.70) >DroSec_CAF1 95904 92 - 1 AGCGGAAAGAAAGAAAUUUACGGCAAGUGCGCGUGCGCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCUAUAAAAAAUCGCAUUAA .((((................(((...((((....))))))).((((((.((.(((......))).)).))))))........))))..... ( -29.80) >DroSim_CAF1 97395 92 - 1 AGCGGAAAUAAAGAAAUUUACGGCAAGUGCGCGUGCGCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCCCUCUAUAAAAAAUCGCAUUAA .((((...((((....)))).(((...((((....))))))).((((.(..(.(((((....))))))..)))))........))))..... ( -25.90) >DroEre_CAF1 98474 90 - 1 AGCGGAAAUA-AGAAAUUUGCGGCAAGUGCGCGUGCGCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCUAUAAA-AAUCGCAUCAA .((((.....-..........(((...((((....))))))).((((((.((.(((......))).)).)))))).....-..))))..... ( -29.80) >consensus AGCGGAAAUAAAGAAAUUUACGGCAAGUGCGCGUGCGCAGCCAGGAGUGCCCUCACACAAAGGUGUGGUCACUCUAUAAAAAAUCGCAUUAA .((((................(((...((((....))))))).((((((.((.(((......))).)).))))))........))))..... (-29.26 = -29.32 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:25 2006