| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,281,165 – 2,281,267 |
| Length | 102 |
| Max. P | 0.805425 |

| Location | 2,281,165 – 2,281,267 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.19 |
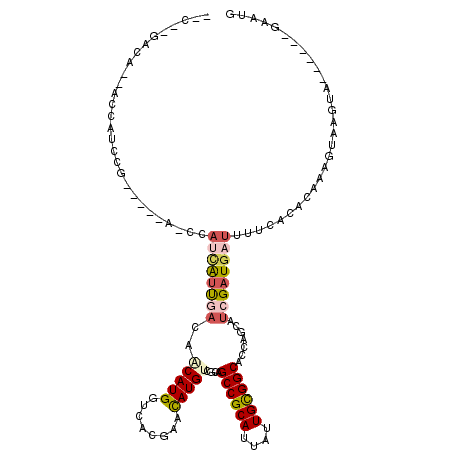
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -16.87 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
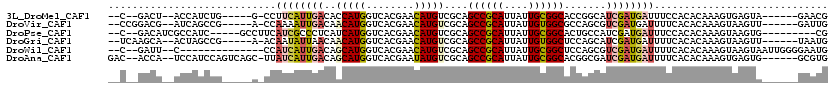
>3L_DroMel_CAF1 2281165 102 + 23771897 --C--GACU--ACCAUCUG-----G-CCUUCAUUGACACCAUGGUCACGAACAUGUCGCAGCCGCAUUAUUGCGGCACCGGCAUCGAUGAUUUCCACACAAAGUGAGUA------GAACG --.--..((--((((((((-----(-((..............))))).....((((((..((((((....))))))..)))))).)))).....(((.....))).)))------).... ( -31.64) >DroVir_CAF1 33403 104 + 1 --CCGGACG--AUCAGCCG-----A-CCAAAAUUGACAACAUGGUCACGAACAUGUCGCAGCCGCAUUAUUGUGGCGCCAGCGUCGAUGAUUUUCACACAAAGUAAGUU------GAUUG --.....((--((((((.(-----(-.....((((((.(((((........))))).((.((((((....))))))....)))))))).....))((.....))..)))------))))) ( -29.90) >DroPse_CAF1 37102 102 + 1 --C--GACAUCGCCAUC-----GCCUUCAUCGCCCUCAUCAUGGUCACGAACAUGUCGCAGCCGCAUUAUUGCGGCACUGCCAUCGAUGAUUUCCACACAAAGUAAGUG---------CG --.--...........(-----(((((........((((((((((..(((.....)))..((((((....))))))...))))).))))).....((.....))))).)---------)) ( -25.30) >DroGri_CAF1 31916 104 + 1 --UCAAGCA--ACUAGCCG-----A-ACAAUAUUAACAACAUGGUCACGAACAUGUCGCAGCCGCAUUAUUGUGGCUCCAGCAUCGAUGAUUUUCACACAAAGUAAGUU------UAAUG --....((.--....)).(-----(-((..............((((((((...(((.(.(((((((....))))))).).)))))).)))))..............)))------).... ( -19.69) >DroWil_CAF1 57685 100 + 1 --C--GAUU--C--------------CCAUCAUUGACAGCAUGGUCACGAACAUGUCGCAGCCGCAUUAUUGCGGCUCCAGCGUCGAUGAUUUUCACACAAAGUAAGUAAUUGGGGAAUG --.--.(((--(--------------(((((((((((.(((((........))))).(((((((((....)))))))...)))))))))))............(((....))))))))). ( -37.80) >DroAna_CAF1 30889 109 + 1 GAC--ACCA--UCCAUCCAGUCAGC-UUAUCAUUGACAGCAUGGUCACGAAUAUGUCGCAGCCGCAUUAUUGCGGCACGGCGAUCGAUGAUUUUCACACAAAGUGAGUG------GCGUG .((--.(((--(((((.(.(((((.-......))))).).))))((((......(((((.((((((....))))))...))))).((......)).......)))))))------).)). ( -34.30) >consensus __C__GACA__ACCAUCCG_____A_CCAUCAUUGACAACAUGGUCACGAACAUGUCGCAGCCGCAUUAUUGCGGCACCAGCAUCGAUGAUUUUCACACAAAGUAAGUA______GAAUG ............................((((((((..(((((........)))))....((((((....)))))).......))))))))............................. (-16.87 = -17.68 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:40 2006