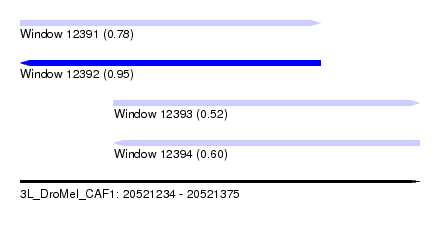
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,521,234 – 20,521,375 |
| Length | 141 |
| Max. P | 0.948501 |

| Location | 20,521,234 – 20,521,340 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -13.88 |
| Energy contribution | -15.86 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20521234 106 + 23771897 AUUGGUUCAUUAGCCCUGCAGUA---GCUAGUAACAAAAGCGUAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCGA-GCGCAAAA--AAAGCAAGGCAGCCGAAAGAGAA .((((((.....(((.(((.((.---.......))....((((.....(((((((.((((...)))).)))))))...-))))....--...))).)))))))))....... ( -29.20) >DroSec_CAF1 63800 112 + 1 AUUGGUUCAUUAGCCCUGCAGUAGCAGCUAGUAAGAAAAGCGGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAAAACGCAAAGAAAGAGCAAGGCAGCCGAAAAAGAA .((((((.....(((.((((((....)))..........(((......(((((((.((((...)))).))))))).....))).........))).)))))))))....... ( -29.70) >DroSim_CAF1 63955 112 + 1 AUUGGUUCAUUAGCCCUGCAGUAGCAGCUAGUAACAAAAGCGGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAAAACGCAAAGAAAGAGCAAGGCAGCCGAAAAAGAA .((((((.....(((.(((.((.((.....)).))....(((......(((((((.((((...)))).))))))).....))).........))).)))))))))....... ( -30.50) >DroEre_CAF1 66296 103 + 1 AUUGGUUCAUUAGCCCUGCAGUA---GCUAGUAACGAAAACAGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAA-ACGGA-----AAAACGAGGCAGCCGAAAUAGAA .((((((.((((((.........---))))))............((..(((((((.((((...)))).)))))))...-.((..-----....))..))))))))....... ( -21.70) >DroAna_CAF1 61308 95 + 1 AUUGGUUCAUUAGCCCUGCAGUA---GCCAGUAACUGAAG------AAAUCCACAGCAAUUAGAUUGGAUUUUGCCAAAA---A-----AAAAAAAAGGAUUGGAAAAAAAA .(..((((..........((((.---.......))))..(------(((((((...(.....)..)))))))).......---.-----........))))..)........ ( -14.10) >consensus AUUGGUUCAUUAGCCCUGCAGUA___GCUAGUAACAAAAGCGGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAAAACGCAAA___AAAGCAAGGCAGCCGAAAAAGAA .(((((((((((((............)))))).......(((......(((((((.((((...)))).))))))).....)))..............).))))))....... (-13.88 = -15.86 + 1.98)

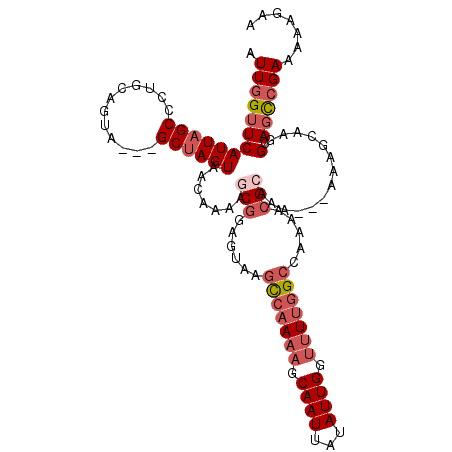

| Location | 20,521,234 – 20,521,340 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -16.94 |
| Energy contribution | -18.06 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20521234 106 - 23771897 UUCUCUUUCGGCUGCCUUGCUUU--UUUUGCGC-UCGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUACGCUUUUGUUACUAGC---UACUGCAGGGCUAAUGAACCAAU ......((((...(((((((...--....(((.-..((((((((.((((...)))).)))))))).....)))....((.....))---....)))))))...))))..... ( -27.60) >DroSec_CAF1 63800 112 - 1 UUCUUUUUCGGCUGCCUUGCUCUUUCUUUGCGUUUUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCCGCUUUUCUUACUAGCUGCUACUGCAGGGCUAAUGAACCAAU ......((((...(((((((.........((((...((((((((.((((...)))).)))))))).))...(((.........))).))....)))))))...))))..... ( -28.32) >DroSim_CAF1 63955 112 - 1 UUCUUUUUCGGCUGCCUUGCUCUUUCUUUGCGUUUUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCCGCUUUUGUUACUAGCUGCUACUGCAGGGCUAAUGAACCAAU ......((((...(((((((.........(((....((((((((.((((...)))).)))))))).....)))....((........))....)))))))...))))..... ( -28.90) >DroEre_CAF1 66296 103 - 1 UUCUAUUUCGGCUGCCUCGUUUU-----UCCGU-UUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCUGUUUUCGUUACUAGC---UACUGCAGGGCUAAUGAACCAAU ......((((...(((.......-----...((-..((((((((.((((...)))).)))))))).)).((((....((.....))---....)))))))...))))..... ( -22.00) >DroAna_CAF1 61308 95 - 1 UUUUUUUUCCAAUCCUUUUUUUU-----U---UUUUGGCAAAAUCCAAUCUAAUUGCUGUGGAUUU------CUUCAGUUACUGGC---UACUGCAGGGCUAAUGAACCAAU ......................(-----(---(.(((((.(((((((..(.....)...)))))))------(((((((.......---.)))).)))))))).)))..... ( -14.60) >consensus UUCUUUUUCGGCUGCCUUGCUUU___UUUGCGUUUUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCCGCUUUUGUUACUAGC___UACUGCAGGGCUAAUGAACCAAU ......((((...(((((((................((((((((.((((...)))).))))))))......(((.........))).......)))))))...))))..... (-16.94 = -18.06 + 1.12)


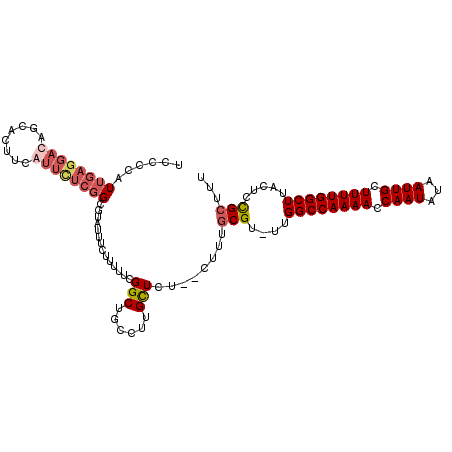
| Location | 20,521,267 – 20,521,375 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -13.86 |
| Energy contribution | -15.86 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20521267 108 + 23771897 AAAGCGUAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCGA-GCGCAAAA--AAAGCAAGGCAGCCGAAAGAGAAAUACGCCGAGAAUAAAGUGCUGUCCUCAAUGGGGA ...((((.....(((((((.((((...)))).)))))))...-))))....--..((((.(((...(....)(....)..)))..........)))).((((.....)))) ( -27.40) >DroSec_CAF1 63836 111 + 1 AAAGCGGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAAAACGCAAAGAAAGAGCAAGGCAGCCGAAAAAGAAAUACGCCGAGAACGAUGUGCUGUCCUCAAUGGGGA ...(((......(((((((.((((...)))).))))))).....))).......(((...(((((((......)....(((.((....)).))))))))))))........ ( -26.10) >DroSim_CAF1 63991 111 + 1 AAAGCGGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAAAACGCAAAGAAAGAGCAAGGCAGCCGAAAAAGAAAUACGCCGAAAACGAAGUGCUGUCCUCAAUGGGGA ...(((......(((((((.((((...)))).))))))).....))).......(((...(((((((.........................).)))))))))........ ( -25.11) >DroEre_CAF1 66329 91 + 1 AAAACAGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAA-ACGGA-----AAAACGAGGCAGCCGAAAUAGAAAUACUCGCAGAAU--------------AAUGGGAG ......(((((.(((((((.((((...)))).)))))))...-.(((.-----............))).........))))).......--------------........ ( -19.12) >consensus AAAGCGGAGUAAGCCAAAAGCAAUUAUAUUGGUUUUGGCCAA_ACGCAAAG__AAAGCAAGGCAGCCGAAAAAGAAAUACGCCGAGAACGAAGUGCUGUCCUCAAUGGGGA ...(((......(((((((.((((...)))).))))))).....))).............((((((............................))))))........... (-13.86 = -15.86 + 2.00)



| Location | 20,521,267 – 20,521,375 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -15.00 |
| Energy contribution | -16.75 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20521267 108 - 23771897 UCCCCAUUGAGGACAGCACUUUAUUCUCGGCGUAUUUCUCUUUCGGCUGCCUUGCUUU--UUUUGCGC-UCGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUACGCUUU ........(((((..........)))))((((((..........(((......)))..--........-..((((((((.((((...)))).))))))))...)))))).. ( -21.80) >DroSec_CAF1 63836 111 - 1 UCCCCAUUGAGGACAGCACAUCGUUCUCGGCGUAUUUCUUUUUCGGCUGCCUUGCUCUUUCUUUGCGUUUUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCCGCUUU ........((((.((((((..((....))..))....(......)))))))))...........(((....((((((((.((((...)))).)))))))).....)))... ( -24.90) >DroSim_CAF1 63991 111 - 1 UCCCCAUUGAGGACAGCACUUCGUUUUCGGCGUAUUUCUUUUUCGGCUGCCUUGCUCUUUCUUUGCGUUUUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCCGCUUU ........((((.((((...(((....)))((...........))))))))))...........(((....((((((((.((((...)))).)))))))).....)))... ( -24.70) >DroEre_CAF1 66329 91 - 1 CUCCCAUU--------------AUUCUGCGAGUAUUUCUAUUUCGGCUGCCUCGUUUU-----UCCGU-UUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCUGUUUU ........--------------.......(((((.........(((..((...))...-----.))).-..((((((((.((((...)))).)))))))))))))...... ( -15.60) >consensus UCCCCAUUGAGGACAGCACUUCAUUCUCGGCGUAUUUCUUUUUCGGCUGCCUUGCUCU__CUUUGCGU_UUGGCCAAAACCAAUAUAAUUGCUUUUGGCUUACUCCGCUUU ......((((((((........))))))))..............(((......)))........(((....((((((((.((((...)))).)))))))).....)))... (-15.00 = -16.75 + 1.75)

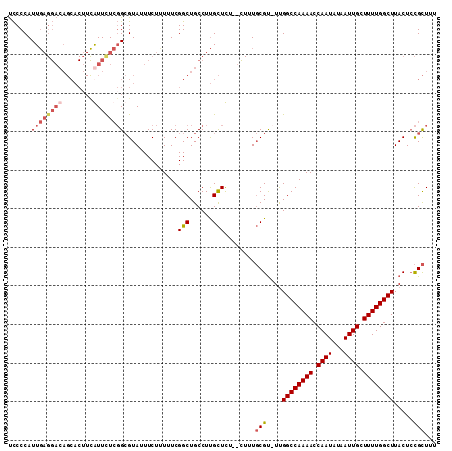
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:52:02 2006