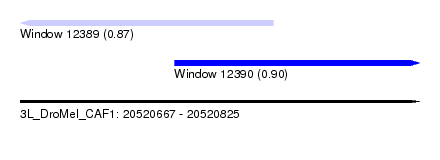
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,520,667 – 20,520,825 |
| Length | 158 |
| Max. P | 0.902434 |

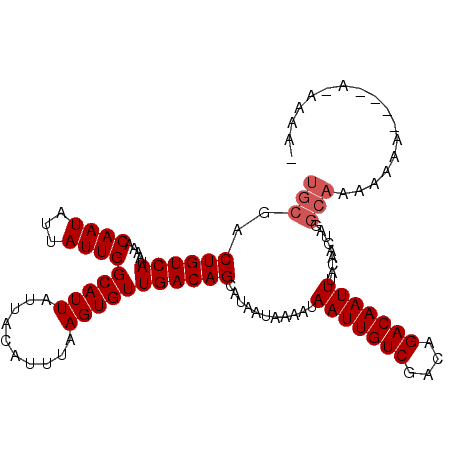
| Location | 20,520,667 – 20,520,767 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -17.08 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20520667 100 - 23771897 UGCGACUGUCAUAAAACAAUAUUAUUGGCAUUAUUACAUUUAAGUGUUGACAGCAUAAUAAAAUAAUUGUCGACAGACAAUUUACAACUAGGCAAAAAAA----------- ((((.((((((.....((((...))))(((((..........))))))))))))..........(((((((....))))))).........)))......----------- ( -17.40) >DroVir_CAF1 70048 106 - 1 UGCGACUGUCAUAAAACAAUAUUAUUGGCAUUAUUACAUUUAAGUGUUGACAGCAUAAUAAAAUAAUUGUCGACAGACAAUUUACAACAACAACAA----CAAAA-ACAAC ...(.((((((.....((((...))))(((((..........))))))))))))..........(((((((....)))))))..............----.....-..... ( -14.50) >DroSec_CAF1 63242 110 - 1 UGCGACUGUCAUAAAACAAUAUUAUUGGCAUUAUUACAUUUAAGUGUUGACAGAAUAAUAAAAUAAUUGUCGACAGACAAUUUACAACUAGGCAAAAAAACAAAA-AAAAA (((..((((((.....((((...))))(((((..........)))))))))))...........(((((((....))))))).........)))...........-..... ( -17.40) >DroSim_CAF1 63388 111 - 1 UGCGACUGUCAUAAAACAAUAUUAUUGGCAUUAUUACAUUUAAGUGUUGACAGCAUAAUAAAAUAAUUGUCGACAGACAAUUUACAACUAGGCAAAAAAACAAAAAAAAAA ((((.((((((.....((((...))))(((((..........))))))))))))..........(((((((....))))))).........)))................. ( -17.40) >DroEre_CAF1 65728 97 - 1 UGCGACUGUCAUAAAACAAUAUUAUUGGCAUUAUUACAUUUAAGUGUUGACAGCAUAAUAAAAUAAUUGUCGACAGACAAUUUACAACUAGGCAAAA-------------- ((((.((((((.....((((...))))(((((..........))))))))))))..........(((((((....))))))).........)))...-------------- ( -17.40) >DroAna_CAF1 60721 106 - 1 UGCGACUGUCAUAAAACAAUAUUAUUGGCAUUAUUACAUUUAAGUGUUGACAGCAUAAUAAAAUAAUUGUCGACAGACAAUUUACAACUAGGCAGGAAAA----A-AAGAU .....(((((...........((((((((......((((....)))).....)).))))))...(((((((....)))))))........))))).....----.-..... ( -18.40) >consensus UGCGACUGUCAUAAAACAAUAUUAUUGGCAUUAUUACAUUUAAGUGUUGACAGCAUAAUAAAAUAAUUGUCGACAGACAAUUUACAACUAGGCAAAAAAA____A_AAAA_ (((..((((((.....((((...))))(((((..........)))))))))))...........(((((((....))))))).........)))................. (-14.90 = -15.40 + 0.50)


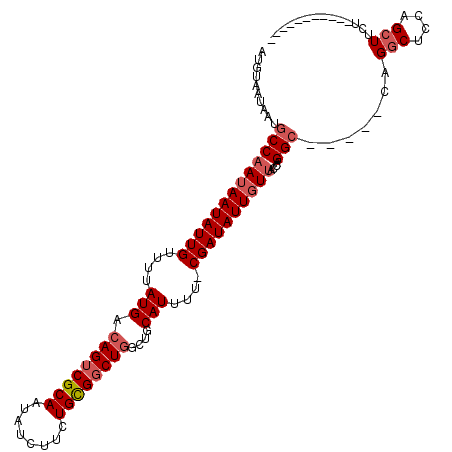
| Location | 20,520,728 – 20,520,825 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.61 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -19.01 |
| Energy contribution | -20.32 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20520728 97 + 23771897 AUGUAAUAAUGCCAAUAAUAUUGUUUUAUGACAGUCGCAAUAUCUUCUGCGGCUGGCUGCAUUUU-CGAUAUUGUUACCAGGC-----CAGGCUCCAGCUUCU----------- ..........(((((((((((((....(((.((((((((........))))))))....)))...-))))))))))....)))-----.((((....))))..----------- ( -27.00) >DroSim_CAF1 63460 108 + 1 AUGUAAUAAUGCCAAUAAUAUUGUUUUAUGACAGUCGCAAUAUCUUCUGCGGCUGGCUGCAUUUU-CGAUAUUGUUACCAGGCCAGGCCAGGCUCCAGCUUCUCAGCUC----- ..........(((((((((((((....(((.((((((((........))))))))....)))...-))))))))))....(((...))).)))...((((....)))).----- ( -32.10) >DroEre_CAF1 65786 112 + 1 AUGUAAUAAUGCCAAUAAUAUUGUUUUAUGACAGUCGCAAUAUCUUCUGCGGCUGGCUGCAUA-U-CGAUAUUGUUACCAGGCCAGGCCAGGCUCCAGCUUUUCAGCUUUUCAG ..........(((((((((((((...((((.((((((((........))))))))....))))-.-))))))))))....(((...))).)))...((((....))))...... ( -33.40) >DroAna_CAF1 60788 96 + 1 AUGUAAUAAUGCCAAUAAUAUUGUUUUAUGACAGUCGCAAUAUCUUCUGUGCCUUGGCGCAUUUUGCGAUAUUUUCGUCCGG-------GGUCUCUGGUUGUU----------- .(((.((((...((((...))))..)))).)))(((((((.......(((((....)))))..))))))).....((.((((-------.....)))).))..----------- ( -19.70) >consensus AUGUAAUAAUGCCAAUAAUAUUGUUUUAUGACAGUCGCAAUAUCUUCUGCGGCUGGCUGCAUUUU_CGAUAUUGUUACCAGGC_____CAGGCUCCAGCUUCU___________ ..........(((((((((((((....(((.((((((((........))))))))....)))....))))))))))....))).......(((....))).............. (-19.01 = -20.32 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:58 2006