
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,491,978 – 20,492,133 |
| Length | 155 |
| Max. P | 0.991205 |

| Location | 20,491,978 – 20,492,075 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.88 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20491978 97 - 23771897 GCAGAUAACAGCACAAAAUGUUGGCAGCCCUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUUUU--GGUGGUUCCA--------UGUGG------------- (((...(((.((.(((((.((....((((.....(((((.(((((((((....))))))))).)))))..)))).)).))))--))).)))...--------)))..------------- ( -24.50) >DroVir_CAF1 43034 89 - 1 GCAGAUAACAGC--AAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGAAUUCUU------GGUGAGG-GA---------GAGA------------- ((........))--..(((((..(((....)))..)))))(((((((((....)))))))))....((((...(((..------...))).-..---------))))------------- ( -20.10) >DroSec_CAF1 34511 98 - 1 GCAGAUAAAAGCACAAAAUGUUGGCAGCCCUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUUUU-CGGUGGUUCCA--------UGGGG------------- ........((((.((((((((..(((....)))..)))))(((((((((....)))))))))......)))))))......(-(.(((....))--------).)).------------- ( -24.40) >DroMoj_CAF1 41897 103 - 1 GCAGAUAACAGC--AAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGCAUUCUA------GGCAAGGCGA---------GGGAGGAGCAGGAGGCU (((((((((.((--..(((((..(((....)))..))))).((.....)))).)))))))....((((((((......------.)))))))).---------.......))........ ( -28.70) >DroAna_CAF1 33034 106 - 1 GCAGAUAACAGCACAAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUUUU-CGGUUGUUCGAACUUCCAAUAGGG------------- ((........))......((((((.(((.(....(((((.(((((((((....))))))))).)))))..))))......((-(((....)))))...))))))...------------- ( -24.10) >DroPer_CAF1 39228 90 - 1 GCAGAUAACAGCACAAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGGUUAUUCUUUUUGGAGG----A-------------------------- ((...(((((........)))))...))(((...(((((.(((((((((....))))))))).)))))...)))..(((((....))))----)-------------------------- ( -20.10) >consensus GCAGAUAACAGCACAAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUUUU__GGUGGUUCGA_________GGGG_____________ ..(((((.........(((((..(((....)))..)))))(((((((((....)))))))))..)))))................................................... (-18.20 = -18.20 + -0.00)



| Location | 20,491,995 – 20,492,113 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -12.23 |
| Energy contribution | -14.15 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20491995 118 + 23771897 AAAAUAAGCCAAGACAUAAUAAAUAACCGCUGUCAUUCAUAAUGUCAACAGGGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCACUCGCGCAAUUCUGGCAACAGAAACAGCGACCAC ......((((..(((((.((.(((.((....)).))).)).))))).....))))............(((((((...(((((.....)))))..((((....)))))))))))..... ( -31.80) >DroVir_CAF1 43047 93 + 1 --AAGAAUUCAAGACAUAAUAAAUAACCGCUGUCAUUCAUAAUGUCAACAGUGCUGCCAACAUUUU--GCUGUUAUCUGCG-------CGCAAUUCUAACAACA-------------- --.(((((((..(((((.((.(((.((....)).))).)).)))))....((((.(..((((....--..))))..).)))-------)).)))))).......-------------- ( -16.00) >DroEre_CAF1 36492 117 + 1 AAAAUAAGCCAAGACAUAAUAAAUAACCGCUGUCAUUCAUAAUGUCAACAGGGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCACUCGCGCAAUUCUGGCAACG-AAACAGCGACCAC ......((((..(((((.((.(((.((....)).))).)).))))).....))))............(((((((.(((((((.....))))).....(....))-))))))))..... ( -26.90) >DroMoj_CAF1 41924 93 + 1 --UAGAAUGCAAGACAUAAUAAAUAACCGCUGUCAUUCAUAAUGUCAACAGUGCUGCCAACAUUUU--GCUGUUAUCUGCG-------CGCAAUUCUAACAACA-------------- --((((((((..(((((.((.(((.((....)).))).)).)))))....((((.(..((((....--..))))..).)))-------))).))))))......-------------- ( -19.30) >DroAna_CAF1 33060 118 + 1 AAAAUAAGCCAAGACAUAAUAAAUAACCGCUGUCAUUCAUAAUGUCAACAGUGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCGCUCGCGCAAUUCUGGCAACAGAAACAGCGACCGC ...........................((((((.............(((((..(............)..)))))...(((((.....))))).(((((....)))))))))))..... ( -30.90) >DroPer_CAF1 39238 92 + 1 AGAAUAACCCAAGACAUAAUAAAUAACCGCUGUCAUUCAUAAUGUCAACAGUGCUGCCAACAUUUUGUGCUGUUAUCUGCUCU--------------------------AGCGACCAC (((.........(((((.((.(((.((....)).))).)).)))))(((((..(............)..))))).)))((...--------------------------.))...... ( -15.00) >consensus AAAAUAAGCCAAGACAUAAUAAAUAACCGCUGUCAUUCAUAAUGUCAACAGUGCUGCCAACAUUUUGUGCUGUUAUCUGCGCC_____CGCAAUUCUGGCAACA_____AGCGACCAC ............(((((.((.(((.((....)).))).)).)))))((((((((............))))))))...(((((.....))))).......................... (-12.23 = -14.15 + 1.92)

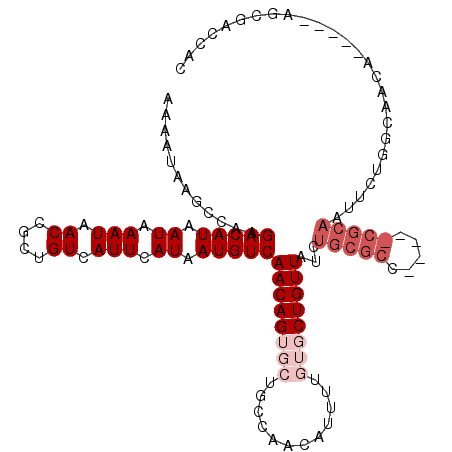
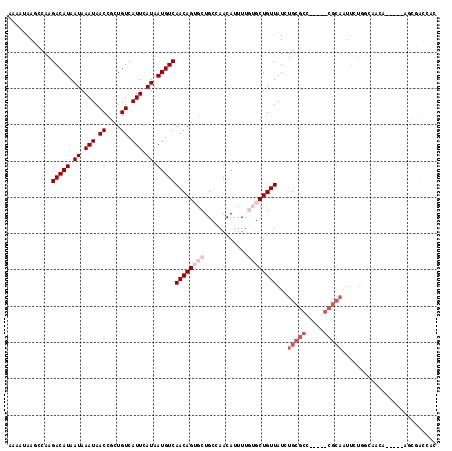
| Location | 20,491,995 – 20,492,113 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -22.72 |
| Energy contribution | -24.47 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20491995 118 - 23771897 GUGGUCGCUGUUUCUGUUGCCAGAAUUGCGCGAGUGGCGCAGAUAACAGCACAAAAUGUUGGCAGCCCUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUU ..(((((((((((((((.(((......((....)))))))))).))))))(((.(((((..(((....)))..)))))(((((((((....)))))))))..)))...))))...... ( -38.50) >DroVir_CAF1 43047 93 - 1 --------------UGUUGUUAGAAUUGCG-------CGCAGAUAACAGC--AAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGAAUUCUU-- --------------((((((((...(((..-------..))).)))))))--).(((((..(((....)))..)))))(((((((((....)))))))))................-- ( -23.90) >DroEre_CAF1 36492 117 - 1 GUGGUCGCUGUUU-CGUUGCCAGAAUUGCGCGAGUGGCGCAGAUAACAGCACAAAAUGUUGGCAGCCCUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUU ..((((.......-.((((((....((((((.....))))))..((((........))))))))))......(((((.(((((((((....))))))))).)))))..))))...... ( -35.60) >DroMoj_CAF1 41924 93 - 1 --------------UGUUGUUAGAAUUGCG-------CGCAGAUAACAGC--AAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGCAUUCUA-- --------------(((((((((..((((.-------...........))--))....))))))))).(((.(((((.(((((((((....))))))))).)))))..))).....-- ( -25.00) >DroAna_CAF1 33060 118 - 1 GCGGUCGCUGUUUCUGUUGCCAGAAUUGCGCGAGCGGCGCAGAUAACAGCACAAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUU ..(((((((((((((((.(((......((....)))))))))).))))))(((.(((((..(((....)))..)))))(((((((((....)))))))))..)))...))))...... ( -40.00) >DroPer_CAF1 39238 92 - 1 GUGGUCGCU--------------------------AGAGCAGAUAACAGCACAAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGGUUAUUCU .((.((...--------------------------.)).))((((((....((((((((..(((....)))..)))))(((((((((....)))))))))......))).)))))).. ( -22.40) >consensus GUGGUCGCU_____UGUUGCCAGAAUUGCG_____GGCGCAGAUAACAGCACAAAAUGUUGGCAGCACUGUUGACAUUAUGAAUGACAGCGGUUAUUUAUUAUGUCUUGGCUUAUUUU ...............((((((....((((((.....))))))..((((........))))))))))......(((((.(((((((((....))))))))).)))))............ (-22.72 = -24.47 + 1.75)



| Location | 20,492,035 – 20,492,133 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20492035 98 + 23771897 AAUGUCAACAGGGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCACUCGCGCAAUUCUGGCAACAGA--AACAGCGACCACAACAACCACAUU----------CUGCACAU ...(((.((((((.((....))))))))((((((...(((((.....)))))..((((....))))--)))))))))...............----------........ ( -26.10) >DroSec_CAF1 34569 100 + 1 AAUGUCAACAGGGCUGCCAACAUUUUGUGCUUUUAUCUGCGCCACUCGCGCAAUUCUGGCAACAGAAGAACAGCGACCACAACAACCAAAUU----------CUGCACAU .((((...((((((.....((.....))((........)))))..((((....(((((....))))).....))))................----------))).)))) ( -22.20) >DroSim_CAF1 34540 100 + 1 AAUGUCAACAGGGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCACUCGCGCAAUUCUGGCAACAGAAGAACAGCGACCACAACAACCACAUU----------CUGCACAU ...(((.((((((.((....))))))))((((((...(((((.....))))).(((((....))))).)))))))))...............----------........ ( -27.20) >DroEre_CAF1 36532 97 + 1 AAUGUCAACAGGGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCACUCGCGCAAUUCUGGCAACG-A--AACAGCGACCACAACAACCACAUU----------CUGCACAU ...(((.((((((.((....))))))))((((((.(((((((.....))))).....(....))-)--)))))))))...............----------........ ( -21.20) >DroAna_CAF1 33100 108 + 1 AAUGUCAACAGUGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCGCUCGCGCAAUUCUGGCAACAGA--AACAGCGACCGCAACAACCACAUUGCGGCGCAUUCGGCGCAU ......(((((..(............)..)))))...(((((((.........(((((....))))--)...(((.((((((........)))))))))...))))))). ( -42.40) >consensus AAUGUCAACAGGGCUGCCAACAUUUUGUGCUGUUAUCUGCGCCACUCGCGCAAUUCUGGCAACAGA__AACAGCGACCACAACAACCACAUU__________CUGCACAU ...(((.((((((.((....))))))))((((((...(((((.....)))))..((((....))))..)))))))))................................. (-22.94 = -23.38 + 0.44)

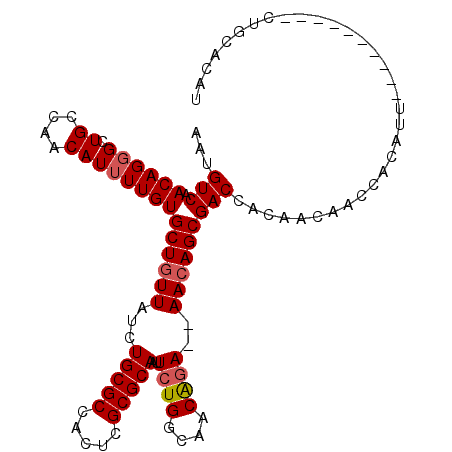

| Location | 20,492,035 – 20,492,133 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -29.10 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20492035 98 - 23771897 AUGUGCAG----------AAUGUGGUUGUUGUGGUCGCUGUU--UCUGUUGCCAGAAUUGCGCGAGUGGCGCAGAUAACAGCACAAAAUGUUGGCAGCCCUGUUGACAUU ........----------..((((.((((((((((.((....--...)).)))....((((((.....))))))))))))))))).(((((..(((....)))..))))) ( -32.00) >DroSec_CAF1 34569 100 - 1 AUGUGCAG----------AAUUUGGUUGUUGUGGUCGCUGUUCUUCUGUUGCCAGAAUUGCGCGAGUGGCGCAGAUAAAAGCACAAAAUGUUGGCAGCCCUGUUGACAUU .(((((..----------.........(((.((((.((.........)).)))).)))(((((.....))))).......))))).(((((..(((....)))..))))) ( -31.50) >DroSim_CAF1 34540 100 - 1 AUGUGCAG----------AAUGUGGUUGUUGUGGUCGCUGUUCUUCUGUUGCCAGAAUUGCGCGAGUGGCGCAGAUAACAGCACAAAAUGUUGGCAGCCCUGUUGACAUU .(((((.(----------((((((..(.....)..))).))))(((((....)))))((((((.....))))))......))))).(((((..(((....)))..))))) ( -32.90) >DroEre_CAF1 36532 97 - 1 AUGUGCAG----------AAUGUGGUUGUUGUGGUCGCUGUU--U-CGUUGCCAGAAUUGCGCGAGUGGCGCAGAUAACAGCACAAAAUGUUGGCAGCCCUGUUGACAUU ........----------..((((.((((((((((.((....--.-.)).)))....((((((.....))))))))))))))))).(((((..(((....)))..))))) ( -31.90) >DroAna_CAF1 33100 108 - 1 AUGCGCCGAAUGCGCCGCAAUGUGGUUGUUGCGGUCGCUGUU--UCUGUUGCCAGAAUUGCGCGAGCGGCGCAGAUAACAGCACAAAAUGUUGGCAGCACUGUUGACAUU .(((((((..(((((((((((......)))))))).((...(--((((....)))))..)))))..))))))).............(((((..(((....)))..))))) ( -46.20) >consensus AUGUGCAG__________AAUGUGGUUGUUGUGGUCGCUGUU__UCUGUUGCCAGAAUUGCGCGAGUGGCGCAGAUAACAGCACAAAAUGUUGGCAGCCCUGUUGACAUU .(((((.....................(((.((((.((.........)).)))).)))(((((.....))))).......))))).(((((..(((....)))..))))) (-29.10 = -29.50 + 0.40)

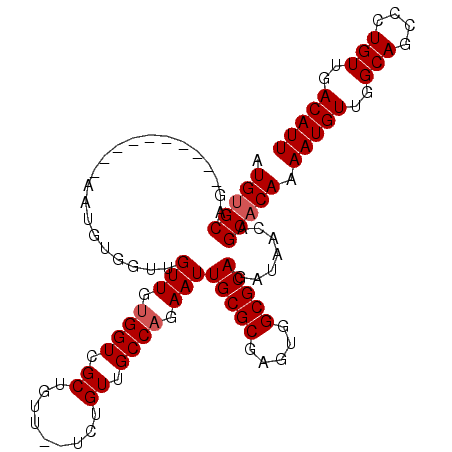
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:45 2006