
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,484,226 – 20,484,470 |
| Length | 244 |
| Max. P | 0.865014 |

| Location | 20,484,226 – 20,484,338 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.47 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20484226 112 - 23771897 UUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGC--GAAAAUUAAUAUCGUCGAUUAAAG------UCU (((((((((.(((.((((........(((((........))))))))).)))(((......((((((...))))))......)))--..............))))))))).------... ( -22.50) >DroPse_CAF1 30626 111 - 1 UUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGGAGCUGCAACUGGCUGCAAAAUGAAUAUCAUCGAUUAAAG--------- (((((((((.(((.((((........(((((........))))))))).)))............((((.((.((((......)))).))...)))).....))))))))).--------- ( -22.50) >DroSim_CAF1 26707 112 - 1 UUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGC--GAAAAUUAAUAUCGUCGAUUAAAG------UCU (((((((((.(((.((((........(((((........))))))))).)))(((......((((((...))))))......)))--..............))))))))).------... ( -22.50) >DroEre_CAF1 28674 118 - 1 UUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGC--GAAAAUUAAUAUCCUCGAUUAAAGUGCAAGUGU ............(((((((..((((.(((((........)))))....(((.(((......((((((...))))))......)))--.)))............))))....))))))).. ( -22.80) >DroAna_CAF1 25320 112 - 1 UUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGACGCAACUGGC--CAAAAUUAAUAACAUCGAUUAAAG------UUU (((((((((.................(((((........)))))....(((((((......((((((...))))))......)))--))))..........))))))))).------... ( -25.80) >DroPer_CAF1 29249 111 - 1 UUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGGAGCUGCAACUGGCUGCAAAAUGAAUAUCAUCGAUUAAAG--------- (((((((((.(((.((((........(((((........))))))))).)))............((((.((.((((......)))).))...)))).....))))))))).--------- ( -22.50) >consensus UUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAACUGGC__CAAAAUUAAUAUCAUCGAUUAAAG______U_U (((((((((.(((.((((........(((((........))))))))).)))(((......((((((...))))))......)))................))))))))).......... (-19.74 = -19.47 + -0.28)

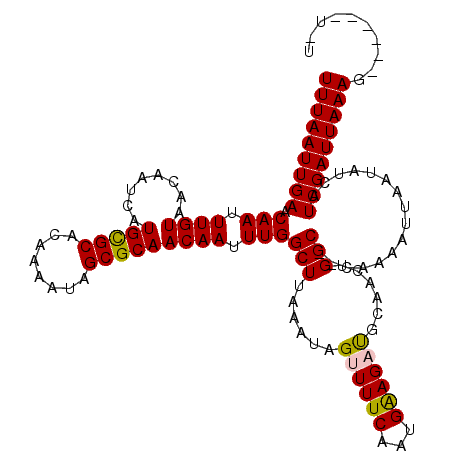

| Location | 20,484,258 – 20,484,376 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.34 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20484258 118 - 23771897 --UCUUGCCUUUCCUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAA --...(((......(((...(((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))).))).))). ( -29.30) >DroPse_CAF1 30657 108 - 1 --ACUCGGCUUUCCUCU----------CCUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGGAGCUGCAA --...(((((((.....----------...((((((((((...(((((.((.....))........(((((........))))).))))).)))))))).))........)))))))... ( -25.09) >DroSec_CAF1 26903 118 - 1 --UCUUGCCUUUUCUAUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAA --...(((.(((((.......((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))))))).))). ( -29.41) >DroEre_CAF1 28712 118 - 1 --UCUUGCCUUUCCUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAA --...(((......(((...(((((((..(..((((((((...(((((.((.....))........(((((........))))).))))).))))))))..)..))))))).))).))). ( -29.30) >DroAna_CAF1 25352 120 - 1 CCUCCUGGCUUUCCUCUGGCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGACGCAA .....(((((.......)))))....((((((((((((((...(((((.((.....))........(((((........))))).))))).)))))).............)))))))).. ( -32.21) >DroPer_CAF1 29280 108 - 1 --ACUCGGCUUUCCUCU----------CCUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGUGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGGAGCUGCAA --...(((((((.....----------...((((((((((...(((((.((.....))........(((((........))))).))))).)))))))).))........)))))))... ( -25.09) >consensus __UCUUGCCUUUCCUCUGCCCAUUGAGCGUCUUUUAAGCCUUUAAUUGAACAAUUUGUAACAAUCAUGCGCACAAAAUAGCGCAACAAUUUGGCUUAAAUAGUUUUCAAUGAAGAUGCAA ..........................((((((((((((((...(((((.((.....))........(((((........))))).))))).)))))).............)))))))).. (-21.42 = -21.86 + 0.44)


| Location | 20,484,376 – 20,484,470 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -33.26 |
| Energy contribution | -34.77 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.865014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20484376 94 + 23771897 CCGGGUUCCAAGACACCCAGAACCC--------CAGAUGCCCAGAUCCCAGGUUCUCCUGACUCCGAUCUGCGGUUCGGAGCUGGGAGUUUCGAGUCUGGGU ..((((((...........))))))--------.....((((((((....((..(((((..((((((........))))))..)))))..))..)))))))) ( -41.00) >DroSec_CAF1 27021 102 + 1 CCGGGUUCCAAGACACCCAGAACCCCCGAUGCCCAGAUGCCCAGAUCCCAGGUUCUACUGGCUCCGAUCUGCGGUCCGGAGCUGGGUGCUUCGAGUCUGGGU ..((((((...........)))))).....(((((((((((((((((((((......))))....)))))).))).((((((.....)))))).)))))))) ( -44.90) >DroSim_CAF1 26857 102 + 1 CCGGGUUCCAAGACACCCAGAACCCCCGAUGCCCAGAUGCCCAGAUCCCAGGUUCUCCUGGCUCCGAUCUGCGGUCCGGAGCUGGGUGCUUCGAGUCUGGGU ..((((((...........)))))).....((((((((((((((((((((((....)))))....)))))).))).((((((.....)))))).)))))))) ( -47.10) >DroEre_CAF1 28830 102 + 1 CCGGGUUCCAAGACACCCAGAGCCCCAGAUGCCCAGAUCCCCAGAUCCCAGGUUCUCCUGGCUCCGAUCUGCAGCGCAGAGCUGGGAGUUUCGAGGCUGGCU ((((((........))))..((((...((.((((.......(((((((((((....)))))....))))))((((.....)))))).)).))..)))))).. ( -37.00) >consensus CCGGGUUCCAAGACACCCAGAACCCCCGAUGCCCAGAUGCCCAGAUCCCAGGUUCUCCUGGCUCCGAUCUGCGGUCCGGAGCUGGGAGCUUCGAGUCUGGGU ((((..((.(((.(((((((..((..............((((((((((((((....)))))....)))))).)))..))..)))))))))).))..)))).. (-33.26 = -34.77 + 1.50)



| Location | 20,484,376 – 20,484,470 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -36.17 |
| Energy contribution | -37.42 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20484376 94 - 23771897 ACCCAGACUCGAAACUCCCAGCUCCGAACCGCAGAUCGGAGUCAGGAGAACCUGGGAUCUGGGCAUCUG--------GGGUUCUGGGUGUCUUGGAACCCGG .((((((((((...((((..(((((((........)))))))..))))....)))).))))))......--------((((((..((...))..)))))).. ( -43.90) >DroSec_CAF1 27021 102 - 1 ACCCAGACUCGAAGCACCCAGCUCCGGACCGCAGAUCGGAGCCAGUAGAACCUGGGAUCUGGGCAUCUGGGCAUCGGGGGUUCUGGGUGUCUUGGAACCCGG ..((.(..((.((((((((((((((((.((.(((((((((.((((......))))..))))...)))))))..))))))...))))))).))).))..).)) ( -41.50) >DroSim_CAF1 26857 102 - 1 ACCCAGACUCGAAGCACCCAGCUCCGGACCGCAGAUCGGAGCCAGGAGAACCUGGGAUCUGGGCAUCUGGGCAUCGGGGGUUCUGGGUGUCUUGGAACCCGG ..((.(..((.((((((((((((((((.((.(((((((((.(((((....)))))..))))...)))))))..))))))...))))))).))).))..).)) ( -44.70) >DroEre_CAF1 28830 102 - 1 AGCCAGCCUCGAAACUCCCAGCUCUGCGCUGCAGAUCGGAGCCAGGAGAACCUGGGAUCUGGGGAUCUGGGCAUCUGGGGCUCUGGGUGUCUUGGAACCCGG ....(((((((.....(((((.(((((...)))))(((((.(((((....)))))..)))))....)))))....)))))))((((((........)))))) ( -42.20) >consensus ACCCAGACUCGAAACACCCAGCUCCGGACCGCAGAUCGGAGCCAGGAGAACCUGGGAUCUGGGCAUCUGGGCAUCGGGGGUUCUGGGUGUCUUGGAACCCGG .((((((((((...((((..(((((((.(....).)))))))..))))....)))).))))))..............((((((..((...))..)))))).. (-36.17 = -37.42 + 1.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:38 2006