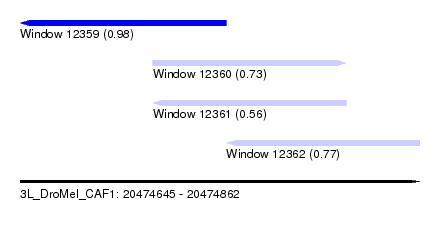
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,474,645 – 20,474,862 |
| Length | 217 |
| Max. P | 0.975020 |

| Location | 20,474,645 – 20,474,757 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20474645 112 - 23771897 CAACGAACGUAUAAGCAUUAUCGCGUAUACGCACUAUAAACAAACAAUUUUACCUUUAUUAAGCAUACGAACAGUGGUACAAGGAUUCAAAAUUCUGAGUGCAAAUAA--------AAUA ........(((((.((......)).)))))(((((...............((((.....................))))..(((((.....))))).)))))......--------.... ( -17.00) >DroSec_CAF1 17466 112 - 1 CAACGAACGUAUACGCAUUAUCGCGUAUACGCACUAUAAACAAACCGUUUCAAUUUUAUAAAGCAUACGAACAGUGGUACAACGAUUCAAAAUUCGGAGUGCAAAUAA--------AAUA .......(((((((((......))))))))).....................(((((((......(((.....)))((((..(((........)))..))))..))))--------))). ( -24.30) >DroSim_CAF1 17342 112 - 1 CAACGAACGUAUACGCAUUAUCGCGUAUACGCACUAUAAACAAACCGUUUCAAUUUUAUAAAGCAUACGAACAGUGGUACAACGAUUCAAAAUUCGGAGUGCAAAUAA--------AAUA .......(((((((((......))))))))).....................(((((((......(((.....)))((((..(((........)))..))))..))))--------))). ( -24.30) >DroEre_CAF1 17265 114 - 1 CAAUGAGCGUAUACGCUUUAUCGCGUAUACGCACUAUAAACAAACCAUUUCACAAUUUU------UACGAACAGUGGUACAAAGAUUCGAAAUCAGGAGUGCAAAUAAUCCUUAAAAAUA ...((((.((((((((......))))))))(((((........((((((((........------...)))..))))).....(((.....)))...)))))........))))...... ( -24.80) >consensus CAACGAACGUAUACGCAUUAUCGCGUAUACGCACUAUAAACAAACCAUUUCAAUUUUAUAAAGCAUACGAACAGUGGUACAACGAUUCAAAAUUCGGAGUGCAAAUAA________AAUA ........((((((((......))))))))(((((..............................(((.....)))......(((........))).))))).................. (-17.85 = -18.60 + 0.75)



| Location | 20,474,717 – 20,474,822 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.89 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20474717 105 + 23771897 UUUAUAGUGCGUAUACGCGAUAAUGCUUAUACGUUCGUUGUGACCUAUCGAUUAUUUCGUGCGAUGUUAUAUCGAUUAACGACUUAUA---------------GCUGUAAAAUGCUGAUA (((((((((((((((.((......)).)))))))..(((((.....((((((.((.((....)).))...))))))..))))).....---------------))))))))......... ( -25.40) >DroSec_CAF1 17538 119 + 1 UUUAUAGUGCGUAUACGCGAUAAUGCGUAUACGUUCGUUGUGACCUAUCGUUUAUUUCAUGCAGCCUUAUAUCGAUUAAC-ACUGUCAGCCGUGGCGUUCUGUGCUGAAAAAUGCUGAUA .((((((.((((((((((......))))))))))...))))))..((((((...((((((((((.........(((....-...))).((....))...))))).)))))...)).)))) ( -28.90) >DroSim_CAF1 17414 119 + 1 UUUAUAGUGCGUAUACGCGAUAAUGCGUAUACGUUCGUUGUGACCUAUCGAUUAUUUUAUGCGACCUUAAAUCGAUUAAC-ACUGUCAGCUGUGGCGUUCGGUGCUGUAAUAUGCUAAUA .((((((..(((((((((......))))))))(..(((..((((..(((((((................)))))))....-...)))....)..)))..).)..)))))).......... ( -29.89) >DroEre_CAF1 17339 119 + 1 UUUAUAGUGCGUAUACGCGAUAAAGCGUAUACGCUCAUUGUGACCUAUCGAUUAUUUCACGCGACGUUAUAUCGAUUAGC-AUCCACAGCUGUGACGUUCUGUGGUGUGAAAUGCUGAUA .(((((((((((((((((......))))))))))..)))))))..(((((..((((((((((.(((..(..(((..((((-.......)))))))..)..))).)))))))))).))))) ( -41.70) >consensus UUUAUAGUGCGUAUACGCGAUAAUGCGUAUACGUUCGUUGUGACCUAUCGAUUAUUUCAUGCGACCUUAUAUCGAUUAAC_ACUGACAGCUGUGGCGUUCUGUGCUGUAAAAUGCUGAUA ((((((((((((((((((......))))))))))..((.((.....((((((..................))))))..)).))....................))))))))......... (-20.58 = -20.89 + 0.31)


| Location | 20,474,717 – 20,474,822 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -16.28 |
| Energy contribution | -17.34 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20474717 105 - 23771897 UAUCAGCAUUUUACAGC---------------UAUAAGUCGUUAAUCGAUAUAACAUCGCACGAAAUAAUCGAUAGGUCACAACGAACGUAUAAGCAUUAUCGCGUAUACGCACUAUAAA ....(((........))---------------).....(((((.((((((......((....))....)))))).(....)))))).((((((.((......)).))))))......... ( -16.30) >DroSec_CAF1 17538 119 - 1 UAUCAGCAUUUUUCAGCACAGAACGCCACGGCUGACAGU-GUUAAUCGAUAUAAGGCUGCAUGAAAUAAACGAUAGGUCACAACGAACGUAUACGCAUUAUCGCGUAUACGCACUAUAAA ((((((((((..(((((...(.....)...))))).)))-)))....))))...((.(((.(((..((.....))..)))........((((((((......)))))))))))))..... ( -25.70) >DroSim_CAF1 17414 119 - 1 UAUUAGCAUAUUACAGCACCGAACGCCACAGCUGACAGU-GUUAAUCGAUUUAAGGUCGCAUAAAAUAAUCGAUAGGUCACAACGAACGUAUACGCAUUAUCGCGUAUACGCACUAUAAA .....((......((((.............))))...((-((..(((((((................)))))))..).))).......((((((((......))))))))))........ ( -26.21) >DroEre_CAF1 17339 119 - 1 UAUCAGCAUUUCACACCACAGAACGUCACAGCUGUGGAU-GCUAAUCGAUAUAACGUCGCGUGAAAUAAUCGAUAGGUCACAAUGAGCGUAUACGCUUUAUCGCGUAUACGCACUAUAAA (((((..(((((((.((((((..........))))))..-......((((.....)))).)))))))..).))))...........((((((((((......))))))))))........ ( -36.10) >consensus UAUCAGCAUUUUACAGCACAGAACGCCACAGCUGACAGU_GUUAAUCGAUAUAACGUCGCAUGAAAUAAUCGAUAGGUCACAACGAACGUAUACGCAUUAUCGCGUAUACGCACUAUAAA (((((((..((((((((.............))))))))..)))...((((.....))))............))))............(((((((((......)))))))))......... (-16.28 = -17.34 + 1.06)

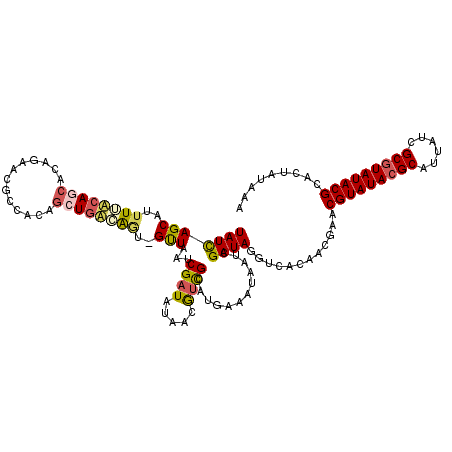
| Location | 20,474,757 – 20,474,862 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20474757 105 - 23771897 AUCCGGUUGCUUAGCAGCUCUGUUAAUUAACAGUGCAGCUUAUCAGCAUUUUACAGC---------------UAUAAGUCGUUAAUCGAUAUAACAUCGCACGAAAUAAUCGAUAGGUCA ....((((((...))))))((((..((((...((((.((((((.(((........))---------------))))))).((((.......))))...))))....))))..)))).... ( -24.90) >DroSec_CAF1 17578 119 - 1 AUUUGGUUGCCUAACUGCUCUGUUAAAUAACAGUGCAGCCUAUCAGCAUUUUUCAGCACAGAACGCCACGGCUGACAGU-GUUAAUCGAUAUAAGGCUGCAUGAAAUAAACGAUAGGUCA ........(((((......(((((....)))))(((((((((((((((((..(((((...(.....)...))))).)))-)))....)))...))))))))............))))).. ( -30.40) >DroSim_CAF1 17454 119 - 1 AUUUGGUUGCCCAACUGCUCUGUUAAAUAACAGUGCAGCCUAUUAGCAUAUUACAGCACCGAACGCCACAGCUGACAGU-GUUAAUCGAUUUAAGGUCGCAUAAAAUAAUCGAUAGGUCA ..((((....))))((((.(((((....))))).))))(((((((((((....((((.............))))...))-)))))((((((................))))))))))... ( -27.71) >DroEre_CAF1 17379 119 - 1 UUUUAGUUUUCUAGCGGCACUGUUAAUUAACAGUGCAGCUUAUCAGCAUUUCACACCACAGAACGUCACAGCUGUGGAU-GCUAAUCGAUAUAACGUCGCGUGAAAUAAUCGAUAGGUCA .....(((....))).((((((((....)))))))).((((((((..(((((((.((((((..........))))))..-......((((.....)))).)))))))..).))))))).. ( -35.10) >consensus AUUUGGUUGCCUAACUGCUCUGUUAAAUAACAGUGCAGCCUAUCAGCAUUUUACAGCACAGAACGCCACAGCUGACAGU_GUUAAUCGAUAUAACGUCGCAUGAAAUAAUCGAUAGGUCA ..((((....))))..((.(((((....))))).)).((((((((((..((((((((.............))))))))..)))...((((.....))))............))))))).. (-18.43 = -18.25 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:32 2006