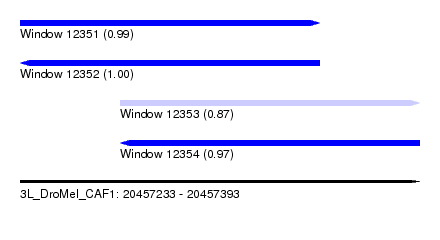
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,457,233 – 20,457,393 |
| Length | 160 |
| Max. P | 0.997953 |

| Location | 20,457,233 – 20,457,353 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -37.69 |
| Consensus MFE | -34.13 |
| Energy contribution | -34.33 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
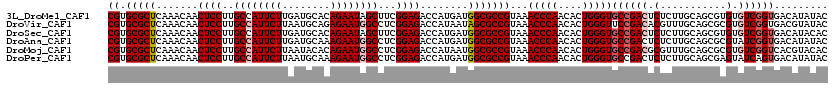
>3L_DroMel_CAF1 20457233 120 + 23771897 UUACUUGGACAGUCAGCCACAGGACAAUGUUGAUGGCUUGCGUGCGCUCAAACAACUCCUUGCCAUUCUUGAUGCACAGAAUAGCUUCGGAGACCAUGAUGGCGCCGUAAACCCAACACU ....((((.......(((((((.......))).))))(((((.(((((.......((((..((.(((((........))))).))...))))........))))))))))..)))).... ( -32.26) >DroVir_CAF1 2835 120 + 1 GCACCUGCACGGUCAGCCAGAGCACAAUGUUGAUGGCUUGCGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUGCAGAGAAUGGCCUCGGAGACCAUAAUAGCGCCGUAAACCCAACACU ((....))..(((..((((.(((.....)))..))))(((((.(((((.......((((..(((((((((......)))))))))...))))........)))))))))))))....... ( -41.46) >DroYak_CAF1 3456 120 + 1 UUACUUGGACAGUCAGCCACAGGACAAUGUUGAUGGCUUGCGUGCGCUCAAACAACUCCUUGCCAUUCUUGAUGCACAGAAUAGCUUCGGAGACCAUGAUGGCGCCGUAAACCCAACACU ....((((.......(((((((.......))).))))(((((.(((((.......((((..((.(((((........))))).))...))))........))))))))))..)))).... ( -32.26) >DroAna_CAF1 2997 120 + 1 GCACUUGGAUCGUCAGCCACAGGACAAUGUUGAUGGCUUGCGUGCGCUCAAACAACUCCUUGCCAUUCUUGAUGCAAAGAAUGGCCUCGGAGACCAUGAUGGCGCCGUAAACCCAACACU ....((((.((((((((...........)))))))).(((((.(((((.......((((..(((((((((......)))))))))...))))........))))))))))..)))).... ( -41.16) >DroMoj_CAF1 2850 120 + 1 GCACCUGCACGGUCAGCCAAAUCACAAUGUUGAUGGCUUGCGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUACACAGAAUGGCCUCGGAGACCAUAAUGGCGCCGUAAACCCAACACU ((....))..((....)).........(((((..((.(((((.(((((.......((((..((((((((........))))))))...))))........)))))))))).))))))).. ( -38.96) >DroPer_CAF1 3067 120 + 1 UCACUUGAAUCGACAGCCACAGGACAAUGUUGAUGGCUUGCGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUGCAAAGAAUGGCCUCGGAGACCAUGAUGGCGCCGUAAACCCAACACU ........(((((((.((...))....)))))))((.(((((.(((((.......((((..(((((((((......)))))))))...))))........)))))))))).))....... ( -40.06) >consensus GCACUUGGACAGUCAGCCACAGGACAAUGUUGAUGGCUUGCGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUGCACAGAAUGGCCUCGGAGACCAUGAUGGCGCCGUAAACCCAACACU ...........................(((((..((.(((((.(((((.......((((..((((((((........))))))))...))))........)))))))))).))))))).. (-34.13 = -34.33 + 0.19)

| Location | 20,457,233 – 20,457,353 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -43.88 |
| Consensus MFE | -41.06 |
| Energy contribution | -40.48 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20457233 120 - 23771897 AGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAAGCUAUUCUGUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACGCAAGCCAUCAACAUUGUCCUGUGGCUGACUGUCCAAGUAA ((((((((((((.((((((((..(..(.((((...((((((((........))))))))..)))))..).)).)))).)).)))))..)))))))....((.(((.....)))))..... ( -41.40) >DroVir_CAF1 2835 120 - 1 AGUGUUGGGUUUACGGCGCUAUUAUGGUCUCCGAGGCCAUUCUCUGCAUUAAGAAUGGCAAGGAGUUGUUUGAGCGCACGCAAGCCAUCAACAUUGUGCUCUGGCUGACCGUGCAGGUGC ((((((((((((.(((((((...(..(.((((...((((((((........))))))))..)))))..)...))))).)).)))))..)))))))(..(.(((((.....)).))))..) ( -47.90) >DroYak_CAF1 3456 120 - 1 AGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAAGCUAUUCUGUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACGCAAGCCAUCAACAUUGUCCUGUGGCUGACUGUCCAAGUAA ((((((((((((.((((((((..(..(.((((...((((((((........))))))))..)))))..).)).)))).)).)))))..)))))))....((.(((.....)))))..... ( -41.40) >DroAna_CAF1 2997 120 - 1 AGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAGGCCAUUCUUUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACGCAAGCCAUCAACAUUGUCCUGUGGCUGACGAUCCAAGUGC ((((((((((((.((((((((..(..(.((((...(((((((((......)))))))))..)))))..).)).)))).)).)))))..)))))))(((........)))........... ( -43.70) >DroMoj_CAF1 2850 120 - 1 AGUGUUGGGUUUACGGCGCCAUUAUGGUCUCCGAGGCCAUUCUGUGUAUUAAGAAUGGCAAGGAGUUGUUUGAGCGCACGCAAGCCAUCAACAUUGUGAUUUGGCUGACCGUGCAGGUGC ((((((((((((.((((((((..(..(.((((...((((((((........))))))))..)))))..).)).)))).)).)))))..)))))))............(((.....))).. ( -45.60) >DroPer_CAF1 3067 120 - 1 AGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAGGCCAUUCUUUGCAUUAAGAAUGGCAAGGAGUUGUUUGAGCGCACGCAAGCCAUCAACAUUGUCCUGUGGCUGUCGAUUCAAGUGA ((((((((((((.((((((((..(..(.((((...(((((((((......)))))))))..)))))..).)).)))).)).)))))..)))))))(((....)))............... ( -43.30) >consensus AGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAGGCCAUUCUGUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACGCAAGCCAUCAACAUUGUCCUGUGGCUGACCGUCCAAGUGA ((((((((((((.((((((((..(..(.((((...((((((((........))))))))..)))))..).)).)))).)).)))))..)))))))........(((.........))).. (-41.06 = -40.48 + -0.58)
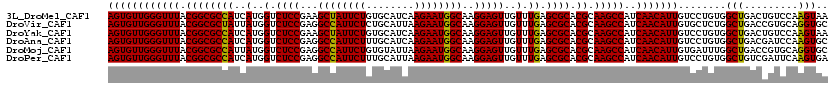

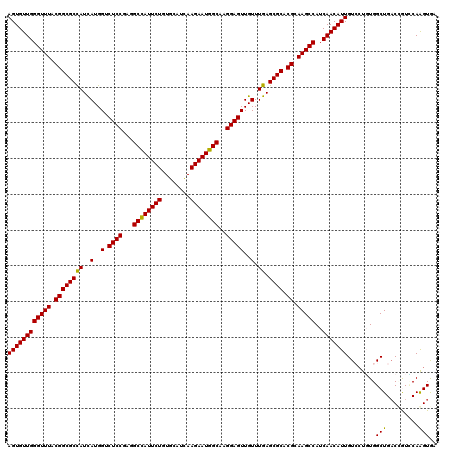
| Location | 20,457,273 – 20,457,393 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -34.56 |
| Energy contribution | -35.43 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20457273 120 + 23771897 CGUGCGCUCAAACAACUCCUUGCCAUUCUUGAUGCACAGAAUAGCUUCGGAGACCAUGAUGGCGCCGUAAACCCAACACUGGGUGCCGACUCUCUUGCAGCGUGUGUCGGUGACAUAUAC ((.(((((.......((((..((.(((((........))))).))...))))........)))))))...(((((....)))))((((((.(.(.......).).))))))......... ( -34.76) >DroVir_CAF1 2875 120 + 1 CGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUGCAGAGAAUGGCCUCGGAGACCAUAAUAGCGCCGUAAACCCAACACUGGGUUCCGACACGUUUGCAGCGCGUGUCGGUGACGUAUAC ((.(((((.......((((..(((((((((......)))))))))...))))........)))))))..((((((....))))))(((((((((.......))))))))).......... ( -47.86) >DroSec_CAF1 3522 120 + 1 CGUGCGCUCAAACAACUCCUUGCCAUUCUUGAUGCACAGAAUAGCUUCGGAGACCAUGAUGGCGCCGUAAACCCAACACUGGGUGCCGACUCUCUUGCAGCGUGUGUCGGUGACAUACAC .(.(((((.......((((..((.(((((........))))).))...))))........))))))(((.(((((....)))))((((((.(.(.......).).))))))....))).. ( -35.26) >DroAna_CAF1 3037 120 + 1 CGUGCGCUCAAACAACUCCUUGCCAUUCUUGAUGCAAAGAAUGGCCUCGGAGACCAUGAUGGCGCCGUAAACCCAACACUGGGUGCCGACUCUCUUGCAGCGCGUAUCGGUGACAUAUAC ...((((((((....((((..(((((((((......)))))))))...)))).......(((((((...............)))))))......))).)))))((((.(....))))).. ( -42.66) >DroMoj_CAF1 2890 120 + 1 CGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUACACAGAAUGGCCUCGGAGACCAUAAUGGCGCCGUAAACCCAACACUGGGUGCCGACGCGUUUGCAGCGCGUGUCGGUCACGUACAC ...((((((((((..((((..((((((((........))))))))...)))).........(((.((...(((((....)))))..)).)))))))).)))))((((((....)).)))) ( -50.00) >DroPer_CAF1 3107 120 + 1 CGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUGCAAAGAAUGGCCUCGGAGACCAUGAUGGCGCCGUAAACCCAACACUGGGUGCCGACUCUCUUGCAGCGAGUAUCAGUGACAUAUAC .((((((........((((..(((((((((......)))))))))...))))....((((((((((...............)))))).((((.(.....).))))))))))).))).... ( -38.36) >consensus CGUGCGCUCAAACAACUCCUUGCCAUUCUUAAUGCACAGAAUGGCCUCGGAGACCAUGAUGGCGCCGUAAACCCAACACUGGGUGCCGACUCUCUUGCAGCGCGUGUCGGUGACAUAUAC ((.(((((.......((((..((((((((........))))))))...))))........)))))))...(((((....)))))((((((.(...........).))))))......... (-34.56 = -35.43 + 0.86)
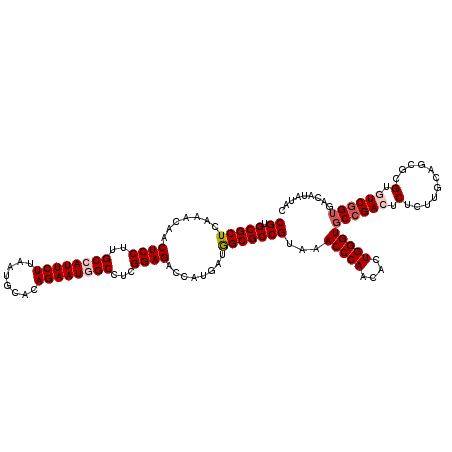
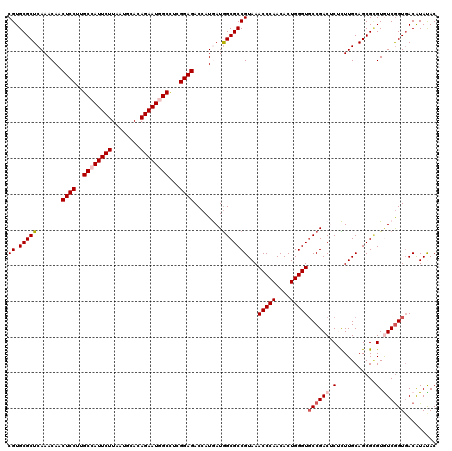
| Location | 20,457,273 – 20,457,393 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -46.08 |
| Consensus MFE | -40.66 |
| Energy contribution | -39.88 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20457273 120 - 23771897 GUAUAUGUCACCGACACACGCUGCAAGAGAGUCGGCACCCAGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAAGCUAUUCUGUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACG ..........(((((((..(((((......).)))).....))))))).....((((((((..(..(.((((...((((((((........))))))))..)))))..).)).)))).)) ( -39.80) >DroVir_CAF1 2875 120 - 1 GUAUACGUCACCGACACGCGCUGCAAACGUGUCGGAACCCAGUGUUGGGUUUACGGCGCUAUUAUGGUCUCCGAGGCCAUUCUCUGCAUUAAGAAUGGCAAGGAGUUGUUUGAGCGCACG .....(((..((((((((.........))))))))(((((......))))).)))(((((...(..(.((((...((((((((........))))))))..)))))..)...)))))... ( -51.20) >DroSec_CAF1 3522 120 - 1 GUGUAUGUCACCGACACACGCUGCAAGAGAGUCGGCACCCAGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAAGCUAUUCUGUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACG ((((.((((...)))).))))(((......((((..((((......))))...))))((((..(..(.((((...((((((((........))))))))..)))))..).)).))))).. ( -41.60) >DroAna_CAF1 3037 120 - 1 GUAUAUGUCACCGAUACGCGCUGCAAGAGAGUCGGCACCCAGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAGGCCAUUCUUUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACG ((((.((....))))))(((((.((((...((((..((((......))))...))))(((.....)))((((...(((((((((......)))))))))..))))...)))))))))... ( -44.40) >DroMoj_CAF1 2890 120 - 1 GUGUACGUGACCGACACGCGCUGCAAACGCGUCGGCACCCAGUGUUGGGUUUACGGCGCCAUUAUGGUCUCCGAGGCCAUUCUGUGUAUUAAGAAUGGCAAGGAGUUGUUUGAGCGCACG ((((.((....))))))(((((.(((((((((((..((((......))))...)))))).........((((...((((((((........))))))))..))))..))))))))))... ( -55.80) >DroPer_CAF1 3107 120 - 1 GUAUAUGUCACUGAUACUCGCUGCAAGAGAGUCGGCACCCAGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAGGCCAUUCUUUGCAUUAAGAAUGGCAAGGAGUUGUUUGAGCGCACG (((.......(((((.(((.......))).))))).((((......)))).))).((((((..(..(.((((...(((((((((......)))))))))..)))))..).)).))))... ( -43.70) >consensus GUAUAUGUCACCGACACGCGCUGCAAGAGAGUCGGCACCCAGUGUUGGGUUUACGGCGCCAUCAUGGUCUCCGAGGCCAUUCUGUGCAUCAAGAAUGGCAAGGAGUUGUUUGAGCGCACG .....(((..(((((((..(((((......).)))).....)))))))....)))((((((..(..(.((((...((((((((........))))))))..)))))..).)).))))... (-40.66 = -39.88 + -0.77)

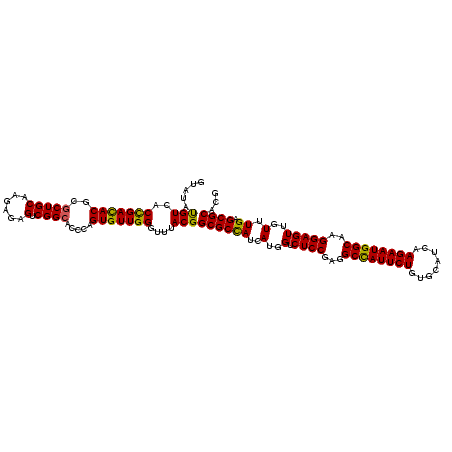
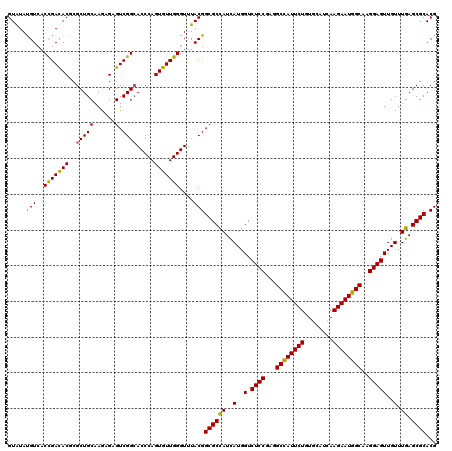
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:23 2006