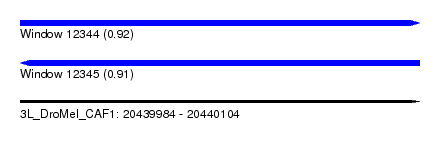
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,439,984 – 20,440,104 |
| Length | 120 |
| Max. P | 0.915224 |

| Location | 20,439,984 – 20,440,104 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -40.06 |
| Consensus MFE | -31.70 |
| Energy contribution | -32.58 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20439984 120 + 23771897 GGGGCCUGCUAACAAUGCCCAUGAUAGCCACUCUAAAGGAGACCUUCCCAGACCACACGUUCCUAAUGAACGGAUUGGUCAUGGGAGUCAAGGGAAUACUCUCGUUUCUGUCGUCGCCCC (((((..((.......))..(((((((..((.........(((..((((((((((..(((((.....)))))...))))).))))))))..((((....))))))..))))))).))))) ( -43.50) >DroSec_CAF1 49685 120 + 1 GGGGCCUGCUGACAAUGCCCAUGAUAGCCACUCUAAAGGAGACCUUCCCAGACCACACGUUCCUCAUGAACGGAUUGGUCAUGGGUGUCAAGGGAAUACUCUCGUUUCUAUCGUUGCCCC (((((..((.......))..(((((((..((.........(((...(((((((((..(((((.....)))))...))))).)))).)))..((((....))))))..))))))).))))) ( -42.10) >DroSim_CAF1 51440 120 + 1 GGGGCCUGCUGACAAUGCCCAUGAUAGCCACUCUAAAGGAGACCUUCCCAGAGCACACGUUCCUCAUGAACGGAUUGGUCAUGGGUGUCAAGGGAAUACUCUCGUUUCUAUCGUCGCCCC (((((....((((...(((((((((..((.((((...((((...)))).)))).....((((.....))))))....))))))))))))).((((....))))............))))) ( -40.30) >DroEre_CAF1 44695 120 + 1 GGGGCUUGCUGACAAUGCCGAUGAUAAGCACUUUAAAAGAAACCUUCCCAGACCACACGUUUCUAAUGAACGGAUUGGUCAUGGGUGUCAAGGGAAUACUAUCGUUUCUAUCGGCGCCCA .(((((((....))).(((((((..((((.............(((((((((((((..(((((.....)))))...))))).))))....))))((......)))))).))))))))))). ( -39.60) >DroYak_CAF1 69612 120 + 1 GGGGCCUUCUGACAAUGCCUAUGAUAGCCACCUUAAAGGAAACCUUCCCCGACCACACGUUCCUAAUGAACGGAUUAGUCAUGGGUGUCAAGGGAAUACUCUCGUUUCUAUCGGCGCCCC ((((((...((((...(((((((((..((........(((.....)))..........((((.....))))))....))))))))))))).((((....)))).........))).))). ( -34.80) >consensus GGGGCCUGCUGACAAUGCCCAUGAUAGCCACUCUAAAGGAGACCUUCCCAGACCACACGUUCCUAAUGAACGGAUUGGUCAUGGGUGUCAAGGGAAUACUCUCGUUUCUAUCGUCGCCCC (((((..((((.((.......)).)))).........((((((...(((((((((..(((((.....)))))...))))).))))......((((....))))))))))......))))) (-31.70 = -32.58 + 0.88)

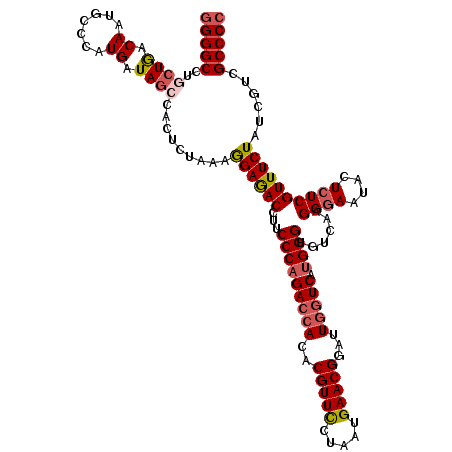

| Location | 20,439,984 – 20,440,104 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -35.74 |
| Energy contribution | -36.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20439984 120 - 23771897 GGGGCGACGACAGAAACGAGAGUAUUCCCUUGACUCCCAUGACCAAUCCGUUCAUUAGGAACGUGUGGUCUGGGAAGGUCUCCUUUAGAGUGGCUAUCAUGGGCAUUGUUAGCAGGCCCC ((((((..(((((..((....))...(((.(((.(((((.(((((...(((((.....)))))..)))))))))).((((.((....).).)))).))).)))..)))))..)..))))) ( -43.60) >DroSec_CAF1 49685 120 - 1 GGGGCAACGAUAGAAACGAGAGUAUUCCCUUGACACCCAUGACCAAUCCGUUCAUGAGGAACGUGUGGUCUGGGAAGGUCUCCUUUAGAGUGGCUAUCAUGGGCAUUGUCAGCAGGCCCC (((((...(((((..((.((((.....((((....((((.(((((...(((((.....)))))..)))))))))))))....))))...))..)))))....((.......))..))))) ( -43.40) >DroSim_CAF1 51440 120 - 1 GGGGCGACGAUAGAAACGAGAGUAUUCCCUUGACACCCAUGACCAAUCCGUUCAUGAGGAACGUGUGCUCUGGGAAGGUCUCCUUUAGAGUGGCUAUCAUGGGCAUUGUCAGCAGGCCCC (((((.......(((((....)).)))..(((((((((((((((.(..(((((.....)))))..)(((((((((.....))))..)))))))...)))))))...))))))...))))) ( -42.50) >DroEre_CAF1 44695 120 - 1 UGGGCGCCGAUAGAAACGAUAGUAUUCCCUUGACACCCAUGACCAAUCCGUUCAUUAGAAACGUGUGGUCUGGGAAGGUUUCUUUUAAAGUGCUUAUCAUCGGCAUUGUCAGCAAGCCCC .((((((((((......(((((((((.....(((.((((.(((((...(((((....).))))..)))))))))...)))........))))).)))))))))).(((....))))))). ( -38.62) >DroYak_CAF1 69612 120 - 1 GGGGCGCCGAUAGAAACGAGAGUAUUCCCUUGACACCCAUGACUAAUCCGUUCAUUAGGAACGUGUGGUCGGGGAAGGUUUCCUUUAAGGUGGCUAUCAUAGGCAUUGUCAGAAGGCCCC (((((((((((((..((....))..((((((((..(((.((((((...(((((.....)))))..))))))))).(((...))))))))).)))))))...))).....(....)))))) ( -40.50) >consensus GGGGCGACGAUAGAAACGAGAGUAUUCCCUUGACACCCAUGACCAAUCCGUUCAUUAGGAACGUGUGGUCUGGGAAGGUCUCCUUUAGAGUGGCUAUCAUGGGCAUUGUCAGCAGGCCCC (((((...(((((..((.((((.....((((....((((.(((((...(((((.....)))))..)))))))))))))....))))...))..)))))...(((...))).....))))) (-35.74 = -36.30 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:14 2006