
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,434,849 – 20,435,083 |
| Length | 234 |
| Max. P | 0.715874 |

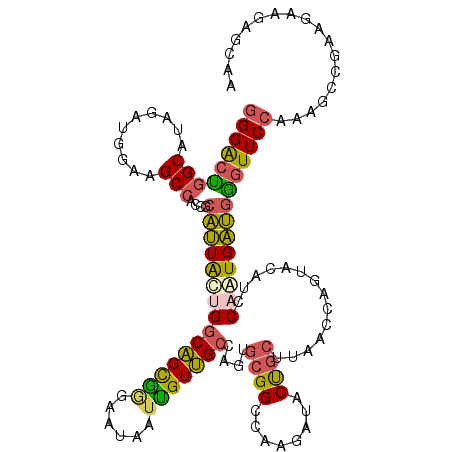
| Location | 20,434,849 – 20,434,969 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -38.13 |
| Consensus MFE | -25.41 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
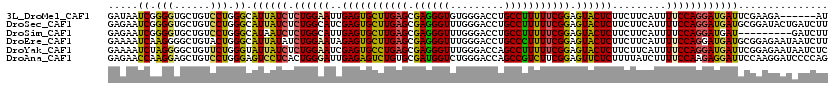
>3L_DroMel_CAF1 20434849 120 - 23771897 GGGACUGGCGUAGAUGGAAGCCACCGCAUUAUUGGCAGCGGGUAUAAUUGUUGCCAGUGCGGCCAAGAUACUGCUUAACCAGUACAUCCAAUGAUGGGUUCCAAAGCCGAAGAAGAACAA .....((((.....(((((.(((((((((...(((((((((......)))))))))))))))......(((((......)))))..........))).)))))..))))........... ( -40.00) >DroSec_CAF1 45068 120 - 1 GGGACUGGCGUAGAUGGAAGCCACCGCAUUAUUGGCAGCGGGUAUAAUUGUUGCCAGUGCGGCCAAGAUACUGCUUAACCAGUACAUCCAAUGAUGGGUUCCAAAGCCGAAGAAAAGCAU .....((((.....(((((.(((((((((...(((((((((......)))))))))))))))......(((((......)))))..........))).)))))..))))........... ( -40.00) >DroSim_CAF1 46740 120 - 1 GGGACUGGCAUAGAUGGAAGCCACCGCAUUAUUGGCAGCGGGAAUAAUUGUUGCCAGUGCGGCCAAGAUACUGCUUAACCAGUACAUCCAAUGAUGGGUUCCACAGCCGAAGAAGAACAA .....((((...(.(((((.(((((((((...(((((((((......)))))))))))))))......(((((......)))))..........))).)))))).))))........... ( -39.00) >DroEre_CAF1 40186 120 - 1 GGGACUGGCAUAGAUGGAAGCAACAGCAUUACAGGCAGUAAAAAUUAUUGUUGCAAAGGCGGCGAAAAUACCGCUUAACCAGUACAUCCAAUGAUGAGUUCCAAAUCCGAGGAAGAGCAA ((((((.((((.((((...(((((((..((((.....))))......)))))))..((((((........))))))........))))..))).).))))))...((....))....... ( -30.20) >DroYak_CAF1 63233 120 - 1 GGGACUGGCAUAGAUGGAAGCCACCGCAUUACUGGCAGCGGGAAUAAUUGUUGCCAUGGCGGCCAAAAUACUGCUUAACCAGUACAUCCAAUGAUGAGUUCCAAAGACGAAGAAUAGCAG ((((((.((((.((((...(((.(((......(((((((((......)))))))))))).))).....(((((......)))))))))..))).).)))))).................. ( -36.40) >DroAna_CAF1 37422 120 - 1 CGGGGCGGCGUAGAGAGAAGCCACCGUGCUGCAGGCCGCCUGGAUGAUGGUGGACACAGAGGCCAGGAUGCCAGCCAACCACAACACCCAGUGGCUGGUUCCAAAGCCAAAGAGGAGGAG ...((((((.(.........(((((((..(.((((...)))).)..)))))))......).))).(((.((((((((..............)))))))))))...)))............ ( -43.20) >consensus GGGACUGGCAUAGAUGGAAGCCACCGCAUUACUGGCAGCGGGAAUAAUUGUUGCCAGUGCGGCCAAGAUACUGCUUAACCAGUACAUCCAAUGAUGGGUUCCAAAGCCGAAGAAGAGCAA (((((((((..........)))....(((((((((((((((......)))))))....((((........))))..............)))))))))))))).................. (-25.41 = -24.92 + -0.49)


| Location | 20,434,969 – 20,435,083 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -39.36 |
| Consensus MFE | -27.55 |
| Energy contribution | -26.45 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20434969 114 + 23771897 GAUAAUCGGGGUGCUGUCCUGGGCAUUAUCUCUGGAAUUGAGUGCUUGAGCGAGGGUGUGGGACCUGCCUUUUUCGGAGUACUCUUCUUCAUUUUCCAGGAUGAUUCGAAGA------AU .....((((((.....))))))(.((((((.((((((..(((((((((((.(((((((.(....))))))))))).)))))))).........)))))))))))).).....------.. ( -34.10) >DroSec_CAF1 45188 120 + 1 GAGAAUCGGGGUGCUGUCCUGGGCAUUAUCUCUGGCAUCGAGUGCUUGAGCGAGGGUUUGGGACCUGCCUUUUUCGGAGUACUCUUCUUCAUUUUCCAGGAUGAUGCGGAUACUGAUCUU ((((.((((.....(((((...((((((((.((((....(((((((((((.((((((..(....).))))))))).))))))))...........))))))))))))))))))))))))) ( -38.36) >DroSim_CAF1 46860 111 + 1 GAGAAUCGGGGUGCUGUCCUGGGCAUAAUCUCUGGCAUUGAGUGCUUGAGCGAGGGUUUGGGACCUGCCUUUUUCGGAGUACUCUUCUUCAUUUUCCAGGAUGAU---------GAUCUU ((((...((((((((((((..(((.....(((.(((((...))))).))).....)))..))))...((......))))))))))((.((((((....)))))).---------)))))) ( -35.30) >DroEre_CAF1 40306 120 + 1 GAAAAUCAAGGGGCUGUACUGGGCAUUAUAUCUGGAAUAGAGUGCUUGAGCGAGGGUUUGGGACCUGCCCUUUUCGGAGUACUCUUCUUCAUUUUCCAGGAUGAUGCGGAGAAUAAUCUU .........(((..(((.((..(((((((.(((((((.((((((((((((.((((((..(....).))))))))).)))))))))........))))))))))))))..)).)))..))) ( -40.70) >DroYak_CAF1 63353 120 + 1 GAAAAUCUAGGGGCUGUUCUGGGUAUUAUCUCUGGAAUCGAGUGCCUGAGCGAGGGUUUGGGACCAGCCUUUUUCGGAGUACUCUUCUUCAUUUUCCAGGAUGAUUCGGAGAAUAAUCUC .........(((..((((((..(.((((((.((((((..(((((((((((.(((((((.(....))))))))))))).)))))).........)))))))))))).)..))))))..))) ( -40.70) >DroAna_CAF1 37542 120 + 1 GAGAACCAAGGAGCUGUCCUGGGAGUCCUCACUGGGAUUGAGAGUCUGUGCGAUGGUCUGGGACCAGCCGUCUUCGGAGUUCUCUUUUAUCUUUUCCAAGAGGAUUCCAAGGAUCCCCAG .........((.(..(((((.(((((((((..(((((..(((((((((...(((((.(((....))))))))..)))).))))).........))))).))))))))).))))).))).. ( -47.00) >consensus GAGAAUCGAGGUGCUGUCCUGGGCAUUAUCUCUGGAAUUGAGUGCUUGAGCGAGGGUUUGGGACCUGCCUUUUUCGGAGUACUCUUCUUCAUUUUCCAGGAUGAUUCGGAGAAUAAUCUU .....((.(((......))).)).((((((.((((((..(((((((((((.((((((.........))))))))))).)))))).........))))))))))))............... (-27.55 = -26.45 + -1.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:10 2006