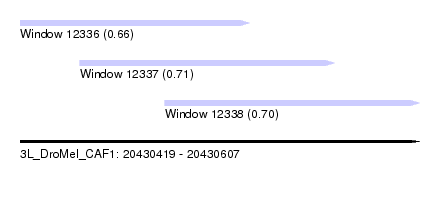
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,430,419 – 20,430,607 |
| Length | 188 |
| Max. P | 0.711511 |

| Location | 20,430,419 – 20,430,527 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -24.34 |
| Energy contribution | -25.95 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
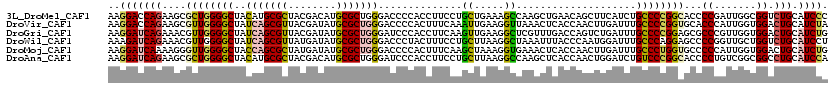
>3L_DroMel_CAF1 20430419 108 + 23771897 UUAUU------------CUCUGCAGACCUAAACGCUAUCAGUUGAGCGAGCUGGUCGACUGGUUCCACCGCCUGAUGCGCAAGGACCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGC .....------------....(((((((....((((........))))....))))..(((((((...(((.....)))...))))))).(((((((......)).)))))......))) ( -34.50) >DroVir_CAF1 36820 120 + 1 UAAUUCAACUUAUCUAAAUUUGCAGUCCCAAGCGCUAUCAGCUGAGCGAGCUGGUCGACUGGUUCCAUCGCCUAAUGCGCAAGGACCAGAAGCGUUGGGGCUAUCAGCGUUACGAUAUGC ..........(((((((..(((.((((((((.(((((((((((.....)))))))...(((((((...(((.....)))...))))))).))))))))))))..)))..))).))))... ( -41.60) >DroGri_CAF1 37590 107 + 1 -------------UUAAAUUUACAGUCCAAAACGUUAUCAGUUGAGCGAGCUGGUCGACUGGUUCCAUCGGCUGAUGCGCAAGGAUCAGAAACGUUGGGGCUAUCAGCGUUACGAUAUGC -------------...........(((...((((((..(((((((.(.....).)))))))((((((.((.(((((.(....).)))))...)).))))))....))))))..))).... ( -35.50) >DroWil_CAF1 42184 108 + 1 AUUUC------------UAUUGCAGGCCCAAACGCUAUCAAUUAAGUGAGCUAGUCGAUUGGUUCCAUCGCCUUAUGCGCAAAGAUCAGAAACGUUGGGGCUAUCAGCGUUAUGAUAUGC ...((------------(.((((.((.((((.((((((((......)))..))).)).))))..))...((.....)))))))))(((..((((((((.....)))))))).)))..... ( -25.40) >DroMoj_CAF1 36317 108 + 1 UAAUU------------AAUUACAGACCCAAGCGAUAUCAGCUGAGCGAGUUGGUUGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAAAAGGGUUGGGGCUACCAGCGCUAUGAUAUGC .....------------..............((.((((((....(((..((((((.....(((((((.(.((((((.(....).)))...)))).)))))))))))))))).)))))))) ( -32.10) >DroAna_CAF1 33046 108 + 1 CAUUC------------CAUUUCAGGCCCAAGCGCUAUCAGCUUAGCGAACUGGUGGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGC .....------------.......(((((.((((((....((...(.((((..(....)..)))))...))(((((.(....).))))).)))))).))))).((((((...)).)))). ( -40.90) >consensus UAAUU____________AAUUGCAGACCCAAACGCUAUCAGCUGAGCGAGCUGGUCGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAGAAGCGUUGGGGCUACCAGCGCUACGAUAUGC ..............................(((((.(((((((.....)))))))....((((((((.((((((((.(....).)))))..))).))))))))...)))))......... (-24.34 = -25.95 + 1.61)

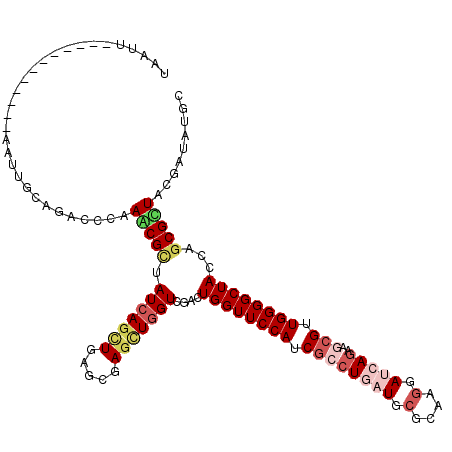
| Location | 20,430,447 – 20,430,567 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -45.75 |
| Consensus MFE | -30.95 |
| Energy contribution | -29.98 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20430447 120 + 23771897 GUUGAGCGAGCUGGUCGACUGGUUCCACCGCCUGAUGCGCAAGGACCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGCUGGGACCCCACCUUCCUGCUGAAAGCCAAGCUGAACAG (((.(((..(((..((..(((((((...(((.....)))...))))))).((((.(((((((.((.((((........)))))))).)))))......)))))).)))...))).))).. ( -44.40) >DroVir_CAF1 36860 120 + 1 GCUGAGCGAGCUGGUCGACUGGUUCCAUCGCCUAAUGCGCAAGGACCAGAAGCGUUGGGGCUAUCAGCGUUACGAUAUGCGCUGGGACCCCACUUUCAAAUUGAAGGUUAAACUCACCAA (((.....)))((((.(((((((((...(((.....)))...)))))))....(((((((...(((((((........)))))))..))))((((((.....))))))..))))))))). ( -42.80) >DroGri_CAF1 37617 120 + 1 GUUGAGCGAGCUGGUCGACUGGUUCCAUCGGCUGAUGCGCAAGGAUCAGAAACGUUGGGGCUAUCAGCGUUACGAUAUGCGCUGGGAUCCCACCUUCAAGUUGAAGGCUCGUUUGACCAG ((..((((((((..(((((((((((((.((.(((((.(....).)))))...)).))))))).(((((((........))))))).............)))))).))))))))..))... ( -56.20) >DroWil_CAF1 42212 120 + 1 AUUAAGUGAGCUAGUCGAUUGGUUCCAUCGCCUUAUGCGCAAAGAUCAGAAACGUUGGGGCUAUCAGCGUUAUGAUAUGCGCUGGGACCCUACUUUCCUGCUUAAGGCUAAAUUUACCCA .....(.(((((((....))))))))...((((((.(((((...((((..((((((((.....)))))))).)))).))))).((((........))))...))))))............ ( -36.40) >DroMoj_CAF1 36345 120 + 1 GCUGAGCGAGUUGGUUGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAAAAGGGUUGGGGCUACCAGCGCUAUGAUAUGCGCUGGGACCCCACUUUCAAGCUAAAGGUGAAACUCACCAA (((((((.((((....)))).)))).......((((.(....).))))..(((((.((((...(((((((........)))))))..)))))))))..)))....((((.....)))).. ( -48.20) >DroAna_CAF1 33074 120 + 1 GCUUAGCGAACUGGUGGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGCUGGGAUCCCACCUUCCUGCUUAAGGCCAAGCUCACCAA ((((((((....(((((..((((((((.((((((((.(....).)))))..))).))))))))((.((((........)))))).....)))))....))))..))))............ ( -46.50) >consensus GCUGAGCGAGCUGGUCGACUGGUUCCAUCGCCUGAUGCGCAAGGAUCAGAAGCGUUGGGGCUACCAGCGCUACGAUAUGCGCUGGGACCCCACCUUCAAGCUGAAGGCUAAACUCACCAA (((.(((.....(((...(((((((...(((.....)))...))))))).......((((...(((((((........)))))))..))))))).....)))...)))............ (-30.95 = -29.98 + -0.97)

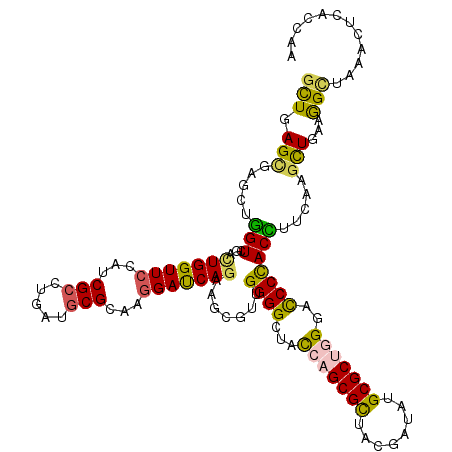
| Location | 20,430,487 – 20,430,607 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -43.41 |
| Consensus MFE | -24.91 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
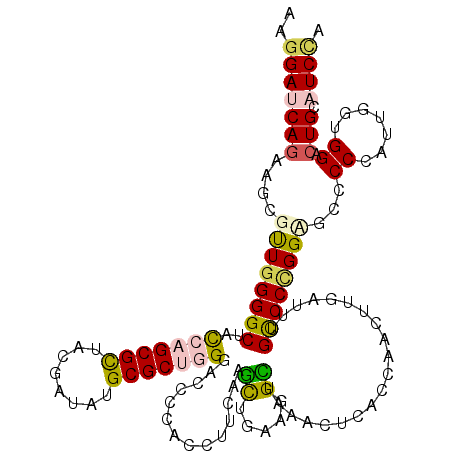
>3L_DroMel_CAF1 20430487 120 + 23771897 AAGGACCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGCUGGGACCCCACCUUCCUGCUGAAAGCCAAGCUGAACAGCUUCAUCUGCCCCGGCACCCCGAUUGGCGGUCUGCAUCCC ..(((((.((((((.(((((((.((.((((........)))))))).))))).)(((..(((........))).)))..)))))....(((.(((....)))...))))))))....... ( -43.60) >DroVir_CAF1 36900 120 + 1 AAGGACCAGAAGCGUUGGGGCUAUCAGCGUUACGAUAUGCGCUGGGACCCCACUUUCAAAUUGAAGGUUAAACUCACCAACUUGAUUUGCCCCGGUGCACCCAUUGGUGGACUGCAUCUA .......(((.((...(((((.((((((((........))))(((((....((((((.....)))))).....)).)))...))))..)))))(((.((((....)))).))))).))). ( -40.20) >DroGri_CAF1 37657 120 + 1 AAGGAUCAGAAACGUUGGGGCUAUCAGCGUUACGAUAUGCGCUGGGAUCCCACCUUCAAGUUGAAGGCUCGUUUGACCAGUCUGAUUUGCCCCGGAGCGCCCGUUGGUGGACUGCAUCUG ..(((((((.....(((((((.((((((((......))))(((((.......(((((.....))))).........))))).))))..)))))))..((((....))))..))).)))). ( -40.29) >DroWil_CAF1 42252 120 + 1 AAAGAUCAGAAACGUUGGGGCUAUCAGCGUUAUGAUAUGCGCUGGGACCCUACUUUCCUGCUUAAGGCUAAAUUUACCCAAUGGAUUUGCCCAGGAGCCCCGGUUGCUGGUCUGCAUCCU ..(((((((.(((..((((((..(((((((........)))))))..........(((((.....(((.(((((((.....)))))))))))))))))))))))).)))))))....... ( -43.70) >DroMoj_CAF1 36385 120 + 1 AAGGAUCAAAAGGGUUGGGGCUACCAGCGCUAUGAUAUGCGCUGGGACCCCACUUUCAAGCUAAAGGUGAAACUCACCAACUUGAUUUGCCCUGGUGCCCCCAUUGGUGGACUGCAUCUG ..........((((((((((...(((((((........)))))))..)))))...(((((.....((((.....))))..)))))...)))))(((((..(((....)))...))))).. ( -46.90) >DroAna_CAF1 33114 120 + 1 AAGGAUCAGAAGCGCUGGGGCUACAUGCGCUACGACAUGCGCUGGGAUCCCACCUUCCUGCUUAAGGCCAAGCUCACCAACUGGAUCUGUCCCGGCACCCCUGUCGGCGGCCUGCAUCCA ..(((((((....((((((((..((.((((........))))))((((((.........((((......)))).........)))))))))))))).((((....)).)).))).)))). ( -45.77) >consensus AAGGAUCAGAAGCGUUGGGGCUACCAGCGCUACGAUAUGCGCUGGGACCCCACCUUCAAGCUGAAGGCUAAACUCACCAACUUGAUUUGCCCCGGAGCCCCCAUUGGUGGACUGCAUCCA ..(((((((....((((((((..(((((((........)))))))..............((.....))....................))))))))...((.......)).))).)))). (-24.91 = -24.92 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:07 2006