
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,423,550 – 20,423,653 |
| Length | 103 |
| Max. P | 0.896797 |

| Location | 20,423,550 – 20,423,653 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -19.45 |
| Energy contribution | -21.62 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
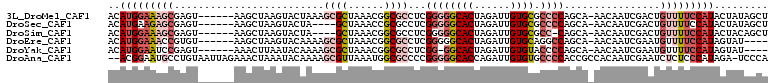
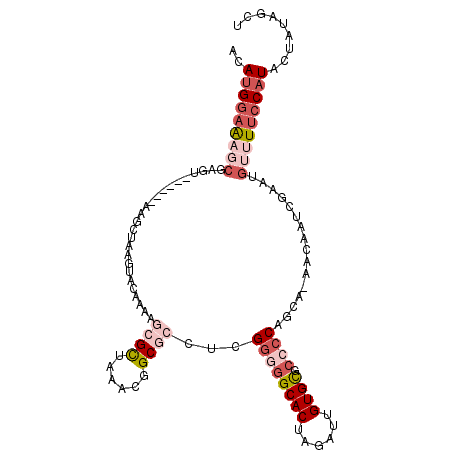
>3L_DroMel_CAF1 20423550 103 + 23771897 ACAUGGAAAGCGAGU------AAGCUAAGUACUAAAGCGCUAAACGGCGCCUCGGGGGCACUAGAUUGUGCGCCCCAGCA-AACAAUCGACUGUUUUCCAUACUAUAGCU ..(((((((((.(((------..(((..........(((((....)))))...((((((((......)))).))))))).-..(....)))))))))))))......... ( -36.40) >DroSec_CAF1 33781 99 + 1 ACAUGAAGAGCGAGU------AAGCUAAGUACUA----GCUAAACCGCGCCUCGGGGGCACUAGAUUGUGCGCCCCAGCA-AACAAUCGACUGUUUUCCAUACUAUAGCU ....((((((((.((------.(((((.....))----)))..)))))((...((((((((......)))).)))).)).-............)))))............ ( -27.60) >DroSim_CAF1 34775 98 + 1 ACAUGGAAAGCGAGU------AAGCUAAGUACUA----GCUAAACGGCGCCUCGGGGGCACUAGAUUGUGCGCC-CAGCA-AACAAUCGACUGUUUUCCAUACUACAGCU ..(((((((((.(((------.(((((.....))----))).......((...(((.((((......)))).))-).)).-........))))))))))))......... ( -32.50) >DroEre_CAF1 29920 99 + 1 ACAUGGAAACCGUGU------AAGCUAAGUACAAAAGCGCUAAACGGCGCCUCGGGGGCACUAGAUUGUGCAGGCCAGCA-AACAAUCGAAUGUUUUCCAUAGUAU---- ((((((...))))))------..((((.........(((((....)))))...(..((((...((((((((......)).-.))))))...))))..)..))))..---- ( -27.30) >DroYak_CAF1 51335 98 + 1 ACAUGGAAUCCGAGU------AAACUUAAUACAAAAGCGCUAAACGGCGCCUCGG-GGCACUAGAUUGUGUACCCCAGCA-AACAAUCGAAUGUUUUCCAUAGUAU---- ((((((((((((((.------...............(((((....))))))))))-)(((...((((((...........-.))))))...))).)))))).))..---- ( -25.49) >DroAna_CAF1 27157 107 + 1 --ACGGAAUGCCUGUAAUUAGAAACUAAAUACAAAAGCGUUAAAUGGCGCCCCGGGGGCACCAGAUUGUGUGCCCCACCGCCACAAUCGAAUCUCUCCCAUAGA-UCCCA --..(((((((.((((.((((...)))).))))...)))))...(((((.....((((((((.....).)))))))..)))))..............)).....-..... ( -31.10) >consensus ACAUGGAAAGCGAGU______AAGCUAAGUACAAAAGCGCUAAACGGCGCCUCGGGGGCACUAGAUUGUGCGCCCCAGCA_AACAAUCGAAUGUUUUCCAUACUAUAGCU ..(((((((((.........................((((......))))...((((((((......)))).))))................)))))))))......... (-19.45 = -21.62 + 2.17)

| Location | 20,423,550 – 20,423,653 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -19.05 |
| Energy contribution | -20.88 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
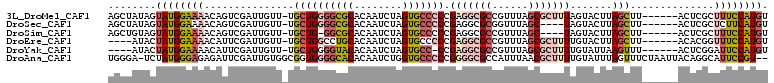
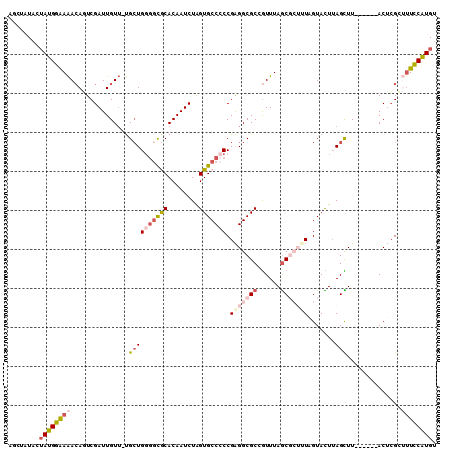
>3L_DroMel_CAF1 20423550 103 - 23771897 AGCUAUAGUAUGGAAAACAGUCGAUUGUU-UGCUGGGGCGCACAAUCUAGUGCCCCCGAGGCGCCGUUUAGCGCUUUAGUACUUAGCUU------ACUCGCUUUCCAUGU ........(((((((...((.(((..((.-.(((((((.((((......))))))))(((((((......))))))).......)))..------)))))))))))))). ( -37.50) >DroSec_CAF1 33781 99 - 1 AGCUAUAGUAUGGAAAACAGUCGAUUGUU-UGCUGGGGCGCACAAUCUAGUGCCCCCGAGGCGCGGUUUAGC----UAGUACUUAGCUU------ACUCGCUCUUCAUGU (((.(((((.((.....))....))))).-.)))((((.((((......))))))))((((.(((((..(((----(((...)))))).------)).))).)))).... ( -34.60) >DroSim_CAF1 34775 98 - 1 AGCUGUAGUAUGGAAAACAGUCGAUUGUU-UGCUG-GGCGCACAAUCUAGUGCCCCCGAGGCGCCGUUUAGC----UAGUACUUAGCUU------ACUCGCUUUCCAUGU ........((((((((((((....)))))-)..((-((.((((......)))).))))(((((..((..(((----(((...)))))).------)).))))))))))). ( -28.40) >DroEre_CAF1 29920 99 - 1 ----AUACUAUGGAAAACAUUCGAUUGUU-UGCUGGCCUGCACAAUCUAGUGCCCCCGAGGCGCCGUUUAGCGCUUUUGUACUUAGCUU------ACACGGUUUCCAUGU ----....((((((((....(((..(((.-.(((((...((((......))))..))(((((((......))))))).......)))..------)))))))))))))). ( -28.40) >DroYak_CAF1 51335 98 - 1 ----AUACUAUGGAAAACAUUCGAUUGUU-UGCUGGGGUACACAAUCUAGUGCC-CCGAGGCGCCGUUUAGCGCUUUUGUAUUAAGUUU------ACUCGGAUUCCAUGU ----....((((((....((((((..((.-.((((((((((........)))))-))(((((((......))))))).......)))..------)))))))))))))). ( -34.40) >DroAna_CAF1 27157 107 - 1 UGGGA-UCUAUGGGAGAGAUUCGAUUGUGGCGGUGGGGCACACAAUCUGGUGCCCCCGGGGCGCCAUUUAACGCUUUUGUAUUUAGUUUCUAAUUACAGGCAUUCCGU-- ..(((-(((.......))))))(((((((((......)).)))))))((((((((...))))))))......(((..((((.((((...)))).))))))).......-- ( -36.70) >consensus AGCUAUACUAUGGAAAACAGUCGAUUGUU_UGCUGGGGCGCACAAUCUAGUGCCCCCGAGGCGCCGUUUAGCGCUUUAGUACUUAGCUU______ACUCGCUUUCCAUGU ........((((((((...............((((((((((........))))))).(((((((......))))))).......)))..............)))))))). (-19.05 = -20.88 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:51:03 2006