| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,398,001 – 20,398,107 |
| Length | 106 |
| Max. P | 0.808421 |

| Location | 20,398,001 – 20,398,107 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.62 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
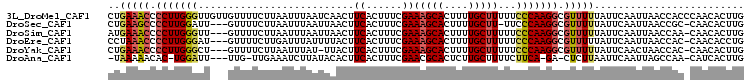
>3L_DroMel_CAF1 20398001 106 + 23771897 CUGAAACCCCUUGGGUUGUUGUUUUCUUAAUUUAAUCAACUUCACUUUCGAAAGCACUUUUGCUUUUUCCCAAGGCGUUUUUAUUCAAUUAACCACCCAACACUUG ..(((((.(((((((..((((...............)))).........(((((((....))))))).))))))).)))))......................... ( -21.86) >DroSec_CAF1 12258 101 + 1 CUGAAGCCCCUUGGAUU---GUUUUCUUAAUUUAAUUAACUUCACUUUCGAAAGCACUUUUGCUU-UUCCCAAGGCGUUUUUAUUCAAUUAACCGC-CAACACUUG .(((((....(((((((---(......))))))))....))))).....(((((((....)))))-)).....((((...(((......))).)))-)........ ( -17.70) >DroSim_CAF1 16509 102 + 1 AUGAAACCCCUUGGGUU---GUUUUCUUAAUUUAAUUAACUUCACUUUCGAAAGCACUUUUGCUUUUUCCCAAGGCGUUUUUAUUCAAUUAACCAA-CAACACUUG ..(((((.(((((((..---(((..............))).........(((((((....))))))).))))))).)))))...............-......... ( -19.14) >DroEre_CAF1 12339 102 + 1 CCUAAACCCCUUGGGAU---GUUUUCUUGAUUUAUUUUACUUCACUUUCGAAAGCACUUUUGCUUUUUCCCAAGGCGUUUUUAUUCAAUUAACCAC-CAACACCUG ...((((.((((((((.---.......(((...........))).....(((((((....))))))))))))))).))))................-......... ( -20.80) >DroYak_CAF1 30909 101 + 1 CUGAAACCCCUUGGGCU---GUUUUCUUAAUUUAU-UUACUUCACUUUCGAAAGCACUUUUGCUUUUUCCCAAGGCGUUUUUAUUCAACUAACCAC-CAACACUUG ..(((((.(((((((..---.......(((.....-)))..........(((((((....))))))).))))))).)))))...............-......... ( -18.60) >DroAna_CAF1 11703 97 + 1 -UAAAAACAC-UGGAUU---UUG-UUGAAAUCUUAUACACUUCACUUUCGAACGCACUCUUGCUUUUCUUCA-GA-CUCUUAAUUCAAUUAGCCAA-CAUCACUUG -.........-(((...---..(-(((((...........(((......))).(((....))).........-..-.......))))))...))).-......... ( -8.40) >consensus CUGAAACCCCUUGGGUU___GUUUUCUUAAUUUAAUUAACUUCACUUUCGAAAGCACUUUUGCUUUUUCCCAAGGCGUUUUUAUUCAAUUAACCAC_CAACACUUG ..(((((.(((((((..........................((......))(((((....)))))...))))))).)))))......................... (-13.18 = -13.62 + 0.43)
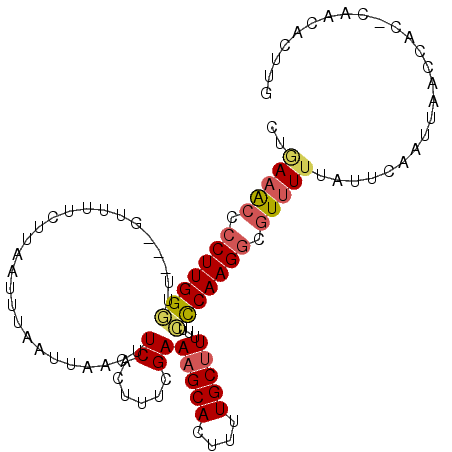
| Location | 20,398,001 – 20,398,107 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.05 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20398001 106 - 23771897 CAAGUGUUGGGUGGUUAAUUGAAUAAAAACGCCUUGGGAAAAAGCAAAAGUGCUUUCGAAAGUGAAGUUGAUUAAAUUAAGAAAACAACAACCCAAGGGGUUUCAG ....((((.(((.....))).)))).((((.(((((((..((((((....))))))..........((((...............))))..))))))).))))... ( -24.76) >DroSec_CAF1 12258 101 - 1 CAAGUGUUG-GCGGUUAAUUGAAUAAAAACGCCUUGGGAA-AAGCAAAAGUGCUUUCGAAAGUGAAGUUAAUUAAAUUAAGAAAAC---AAUCCAAGGGGCUUCAG .((((.(((-(((.(((......)))...))))(((((.(-(((((....))))))(....)........................---..))))))).))))... ( -16.90) >DroSim_CAF1 16509 102 - 1 CAAGUGUUG-UUGGUUAAUUGAAUAAAAACGCCUUGGGAAAAAGCAAAAGUGCUUUCGAAAGUGAAGUUAAUUAAAUUAAGAAAAC---AACCCAAGGGGUUUCAU ......(((-((.(.....).)))))((((.(((((((..((((((....))))))(....)........................---..))))))).))))... ( -22.50) >DroEre_CAF1 12339 102 - 1 CAGGUGUUG-GUGGUUAAUUGAAUAAAAACGCCUUGGGAAAAAGCAAAAGUGCUUUCGAAAGUGAAGUAAAAUAAAUCAAGAAAAC---AUCCCAAGGGGUUUAGG ......(..-((.....))..)....((((.((((((((.((((((....))))))(....)........................---.)))))))).))))... ( -24.50) >DroYak_CAF1 30909 101 - 1 CAAGUGUUG-GUGGUUAGUUGAAUAAAAACGCCUUGGGAAAAAGCAAAAGUGCUUUCGAAAGUGAAGUAA-AUAAAUUAAGAAAAC---AGCCCAAGGGGUUUCAG (((.((...-.....)).))).....((((.(((((((..((((((....))))))(....)........-...............---..))))))).))))... ( -22.60) >DroAna_CAF1 11703 97 - 1 CAAGUGAUG-UUGGCUAAUUGAAUUAAGAG-UC-UGAAGAAAAGCAAGAGUGCGUUCGAAAGUGAAGUGUAUAAGAUUUCAA-CAA---AAUCCA-GUGUUUUUA- .....((..-((((....((((((......-((-....))...(((....)))))))))...((((((.......)))))).-...---...)))-)..))....- ( -12.20) >consensus CAAGUGUUG_GUGGUUAAUUGAAUAAAAACGCCUUGGGAAAAAGCAAAAGUGCUUUCGAAAGUGAAGUUAAUUAAAUUAAGAAAAC___AACCCAAGGGGUUUCAG ..........................((((.(((((((..((((((....))))))(....).............................))))))).))))... (-16.95 = -17.05 + 0.10)


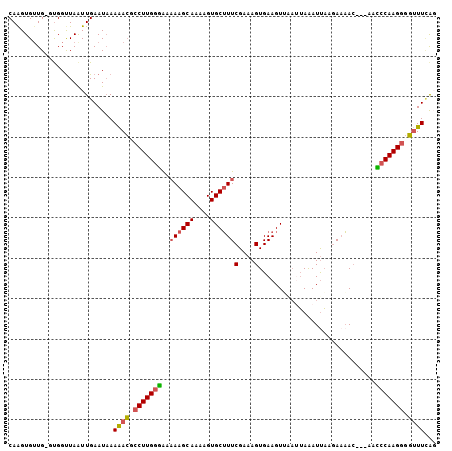
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:59 2006