
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,389,657 – 20,389,763 |
| Length | 106 |
| Max. P | 0.564774 |

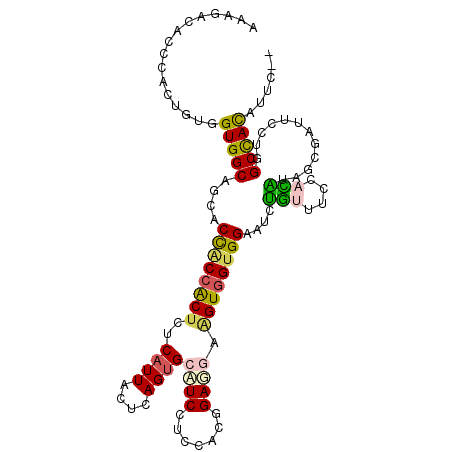
| Location | 20,389,657 – 20,389,763 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -17.10 |
| Energy contribution | -16.27 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20389657 106 + 23771897 AAAGACACCCACUGUGGUGGCAGCGCUACCACUCUCAUUACUCAGUGCAUCCUCCAUGGAUGAAGUUGUGGAAUCUGUAUCCACAUAACGAUUCCUGGCUAUGUUU-- ..(((((......((((((((...)))))))).........((((..(((((.....)))))....((((((.......)))))).........))))...)))))-- ( -29.00) >DroPse_CAF1 4806 106 + 1 AAAAACUCCGACUGUUGUGGCCGACCCGCCGCUCUCAUUGCUCAGUGUCUCCUCGGCCGAGUUGGUGGUGGUGUCCACUUCCAUGUAGCGGUUUCUGGCCACAUUC-- ...............((((((((....((((((..((..((((.(.(((.....))))))))(((.(((((...))))).))))).))))))...))))))))...-- ( -36.80) >DroSim_CAF1 8191 106 + 1 AAAGACACCCACUGUGGUGGCGGCACUACCACUCUCAUUACUCAGUGCAUCCUCCAUGGAUGAAGUGGUGGAAUCUGUGUCCACAUAGCGAUUCCUGGCUAUGUUU-- ...(((((.....(((((((.....)))))))((.(((((((.....(((((.....))))).)))))))))....))))).(((((((........)))))))..-- ( -37.10) >DroEre_CAF1 4126 106 + 1 AAAGACACCCACUGUGGUGGCAGCACUCCCACUCUCAUUACUCAGUGCAUCCUCCAUGGACGAAGUGGUGGAAUCUGUGUCCACAUAACGAUUCCUGGCUAUGUUC-- ...........((((((.((..(((((................)))))..)).))))))(((.(((.(.((((((((((....))))..))))))).))).)))..-- ( -27.69) >DroAna_CAF1 3720 108 + 1 GAAUACACCUACUGUGGUGGCUGCUCCACCGCUCUCAUUACUCAGAGCAUCUUCCACUGAGGACGUGGUGGAGUCUGUUUCCAUGUAUCGGUUUGUUGCCACAUUUUU .((((.((((((.((((..((.(((((((((((((........))))).(((((....)))))...))))))))..))..)))))))..))).))))........... ( -37.10) >DroPer_CAF1 4614 106 + 1 AAAAACUCCGACUGUUGUGGCCGACCCGCCGCUCUCAUUGCUCAGUGUCUCCUCGGCCGAGUUGGUGGUGGUGUCCACUUCCAUGUAGCGGUUUCUGGCCACAUUC-- ...............((((((((....((((((..((..((((.(.(((.....))))))))(((.(((((...))))).))))).))))))...))))))))...-- ( -36.80) >consensus AAAGACACCCACUGUGGUGGCAGCACCACCACUCUCAUUACUCAGUGCAUCCUCCACGGAGGAAGUGGUGGAAUCUGUUUCCACAUAGCGAUUCCUGGCCACAUUC__ ................(((((....((((((((..((((....))))((((.......)))).))))))))....(((....)))............)))))...... (-17.10 = -16.27 + -0.83)


| Location | 20,389,657 – 20,389,763 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -18.85 |
| Energy contribution | -17.05 |
| Covariance contribution | -1.80 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20389657 106 - 23771897 --AAACAUAGCCAGGAAUCGUUAUGUGGAUACAGAUUCCACAACUUCAUCCAUGGAGGAUGCACUGAGUAAUGAGAGUGGUAGCGCUGCCACCACAGUGGGUGUCUUU --..(((((((........)))))))((((((.....((((.(((((((((.....)))))....))))..((.(.((((((....))))))).)))))))))))).. ( -30.70) >DroPse_CAF1 4806 106 - 1 --GAAUGUGGCCAGAAACCGCUACAUGGAAGUGGACACCACCACCAACUCGGCCGAGGAGACACUGAGCAAUGAGAGCGGCGGGUCGGCCACAACAGUCGGAGUUUUU --...(((((((.((..((((..(.(((..((((...))))..))).(((.((..((......))..))...))).)..)))).)))))))))............... ( -35.80) >DroSim_CAF1 8191 106 - 1 --AAACAUAGCCAGGAAUCGCUAUGUGGACACAGAUUCCACCACUUCAUCCAUGGAGGAUGCACUGAGUAAUGAGAGUGGUAGUGCCGCCACCACAGUGGGUGUCUUU --..(((((((........)))))))((((((.......((((((.(((((.....)))))..((.(....).)))))))).......((((....)))))))))).. ( -34.60) >DroEre_CAF1 4126 106 - 1 --GAACAUAGCCAGGAAUCGUUAUGUGGACACAGAUUCCACCACUUCGUCCAUGGAGGAUGCACUGAGUAAUGAGAGUGGGAGUGCUGCCACCACAGUGGGUGUCUUU --..(((((((........)))))))((((((((.((((((.....(((((.....)))))..((.(....).)).))))))...)).((((....)))))))))).. ( -32.20) >DroAna_CAF1 3720 108 - 1 AAAAAUGUGGCAACAAACCGAUACAUGGAAACAGACUCCACCACGUCCUCAGUGGAAGAUGCUCUGAGUAAUGAGAGCGGUGGAGCAGCCACCACAGUAGGUGUAUUC ......(((((..............((....))(.(((((((...(((.....)))....(((((.(....).))))))))))))).)))))(((.....)))..... ( -38.60) >DroPer_CAF1 4614 106 - 1 --GAAUGUGGCCAGAAACCGCUACAUGGAAGUGGACACCACCACCAACUCGGCCGAGGAGACACUGAGCAAUGAGAGCGGCGGGUCGGCCACAACAGUCGGAGUUUUU --...(((((((.((..((((..(.(((..((((...))))..))).(((.((..((......))..))...))).)..)))).)))))))))............... ( -35.80) >consensus __AAACAUAGCCAGAAACCGCUACAUGGAAACAGACUCCACCACUUCCUCCAUGGAGGAUGCACUGAGUAAUGAGAGCGGUAGGGCGGCCACCACAGUGGGUGUCUUU ....(((((((........)))))))......((((.((....((((.......)))).(((.((.(....).))...(((......)))......))))).)))).. (-18.85 = -17.05 + -1.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:57 2006