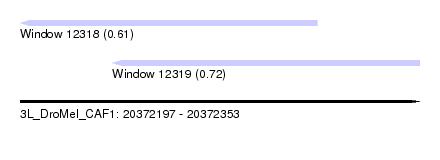
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,372,197 – 20,372,353 |
| Length | 156 |
| Max. P | 0.715179 |

| Location | 20,372,197 – 20,372,313 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -20.42 |
| Energy contribution | -19.62 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
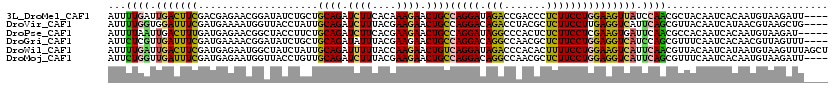
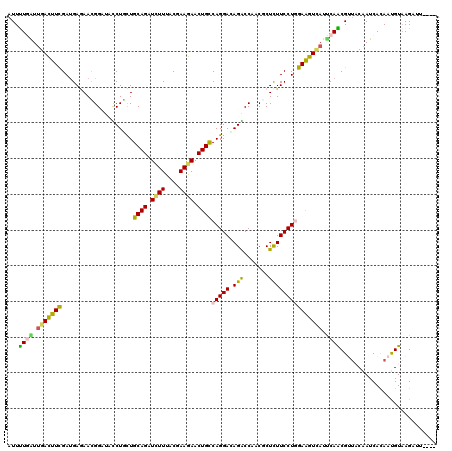
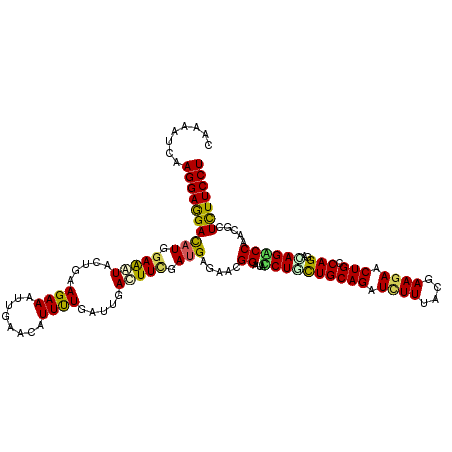
>3L_DroMel_CAF1 20372197 116 - 23771897 AUUUUGAUUGACUUCGACGAGAACGGAUAUCUGCUGCAGAUCUUCACAAAGAACUGCCAGGAUAGACCGACCCUCUUCCUGGAAGUUAUCCAACGCUACAAUCACAAUGUAAGAUU---- (((.((((((.....((.(((..(((.(((((...((((.((((....)))).))))..)))))..)))...))).)).((((.....))))......)))))).)))........---- ( -29.00) >DroVir_CAF1 1694 116 - 1 AUUUUGGUGGAUUUCGAUGAAAAUGGUUACCUAUUGCAGAUCUUUACGAAGAACUGCCAGGACAGACCUACGCUCUUCCUUGAGGUCAUUCAGCGUUACAAUCAUAACGUAAGCUG---- .........((((((((.(((...((((.(((...((((.((((....)))).)))).)))...)))).......))).))))))))...((((.((((.........))))))))---- ( -27.10) >DroPse_CAF1 396 115 - 1 AUUUUAAUUGACUUUGAUGAGAACGGCUACCUUCUGCAGAUCUUCACGAAGAACUGCCAGGAUAGGCCCACUCUCUUCCUCGAAGUGAUUCAACGCCACAAUCACAAUGUAAGAU----- (((((((((((((((((.((((..((((...((((((((.((((....)))).))).)))))..))))...))))....)))))))((((.........)))).)))).))))))----- ( -30.00) >DroGri_CAF1 1654 116 - 1 AUUCUCGUUGAUUUCGAUGAAAACGGAUAUCUGCUGCAGAUAUUUACGAAGAACUGCCAGGACAGGCCAACGCUCUUCCUGGAGGUCAUCCAGCGUUUCAAUCACAACGUUAGUUU---- ....((((((....))))))(((((((((((((...))))))........((.((.((((((..(((....)))..)))))))).)).)))((((((........)))))).))))---- ( -32.60) >DroWil_CAF1 2005 120 - 1 AUUUUGAUUGACUUCGAUGAGAAUGGCUAUCUAUUGCAGAUUUUUACCAAGAACUGUCAGGAUAGACCCACACUUUUCCUGGAAGUCAUUCAACGUUACAAUCAUAAUGUAAGUUUAGCU ....(((.(((((((.(.((((((((((((((...((((.((((....)))).))))..))))))..)))...))))).).))))))).)))((((((......)))))).......... ( -30.00) >DroMoj_CAF1 1765 116 - 1 AUUCUGGUUGAUUUCGAUGAGAAUGGUUACCUGUUGCAGAUCUUUACGAAGAACUGCCAGGACAGGCCAACGCUCUUCCUGGAGGUCAUUCAGCGUUUCAAUCACAAUGUAAGAUU---- (((.(((((((...((....(((((...((((...((((.((((....)))).))))(((((..(((....)))..))))).)))))))))..))..))))))).)))........---- ( -34.10) >consensus AUUUUGAUUGACUUCGAUGAGAACGGAUACCUGCUGCAGAUCUUUACGAAGAACUGCCAGGACAGACCAACGCUCUUCCUGGAAGUCAUUCAACGUUACAAUCACAAUGUAAGAUU____ ...((((.(((((((....................((((.((((....)))).))))(((((.(((.......))))))))))))))).))))........................... (-20.42 = -19.62 + -0.80)
| Location | 20,372,233 – 20,372,353 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -21.50 |
| Energy contribution | -20.73 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
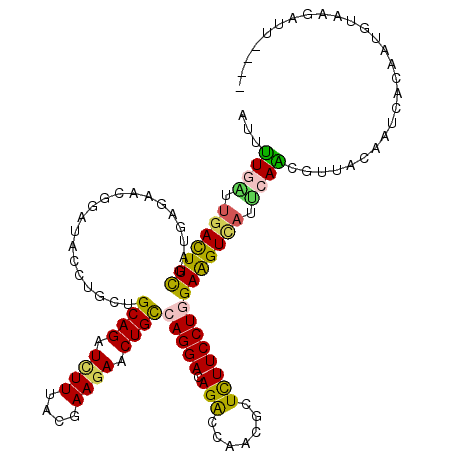
>3L_DroMel_CAF1 20372233 120 - 23771897 CAAGAUCAAGGAGGACAUGGAGGUCUUGAAGAAGUUGAACAUUUUGAUUGACUUCGACGAGAACGGAUAUCUGCUGCAGAUCUUCACAAAGAACUGCCAGGAUAGACCGACCCUCUUCCU ...........((((...((((((((((..(((((..(.(.....).)..)))))..))))).(((.(((((...((((.((((....)))).))))..)))))..)))..))))))))) ( -40.20) >DroVir_CAF1 1730 120 - 1 CAAAAUCAAGGAGGACAUGGAAAUAUUGAAGAAACUGAACAUUUUGGUGGAUUUCGAUGAAAAUGGUUACCUAUUGCAGAUCUUUACGAAGAACUGCCAGGACAGACCUACGCUCUUCCU ........((((((((.......((((((((..((..(.....)..))...)))))))).....((((.(((...((((.((((....)))).)))).)))...))))...).))))))) ( -29.00) >DroPse_CAF1 431 120 - 1 CACAAUCAAGGAGGACAUGGAAAUACUACAGAAAUUGAAUAUUUUAAUUGACUUUGAUGAGAACGGCUACCUUCUGCAGAUCUUCACGAAGAACUGCCAGGAUAGGCCCACUCUCUUCCU ........(((((((.............((((((((((.....))))))...))))..(((...((((...((((((((.((((....)))).))).)))))..))))..)))))))))) ( -28.70) >DroGri_CAF1 1690 120 - 1 UAAAAUUAAGGAGGAUAUGGAAAUAUUAAAGAAAUUGAAUAUUCUCGUUGAUUUCGAUGAAAACGGAUAUCUGCUGCAGAUAUUUACGAAGAACUGCCAGGACAGGCCAACGCUCUUCCU ........(((((((...(..((((((.((....)).))))))..)((((.(((((.(....).(((((((((...))))))))).)))))....(((......)))))))..))))))) ( -28.90) >DroWil_CAF1 2045 120 - 1 AAAGAUCAAGGAAGACAUGAAAAUCCUUCAGAAGUUAAACAUUUUGAUUGACUUCGAUGAGAAUGGCUAUCUAUUGCAGAUUUUUACCAAGAACUGUCAGGAUAGACCCACACUUUUCCU ........(((((((..........(((..((((((((.(.....).))))))))...)))...((((((((...((((.((((....)))).))))..)))))).)).....))))))) ( -26.60) >DroMoj_CAF1 1801 120 - 1 UAAAAUUAAGGAGGAUAUGGAGAUACUGAAGAAACUGAACAUUCUGGUUGAUUUCGAUGAGAAUGGUUACCUGUUGCAGAUCUUUACGAAGAACUGCCAGGACAGGCCAACGCUCUUCCU ........(((((((..(((.....(((...........((((((.((((....)))).))))))....((((..((((.((((....)))).)))))))).))).)))....))))))) ( -37.60) >consensus CAAAAUCAAGGAGGACAUGGAAAUACUGAAGAAAUUGAACAUUUUGAUUGACUUCGAUGAGAACGGAUACCUGCUGCAGAUCUUUACGAAGAACUGCCAGGACAGACCAACGCUCUUCCU ........((((((((((.(((((.....((((........)))).....))))).))).....((...((((((((((.((((....)))).))).)))..)))))).....))))))) (-21.50 = -20.73 + -0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:48 2006