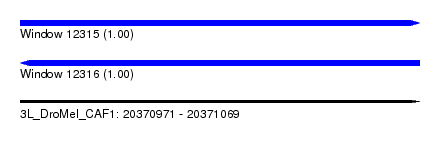
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,370,971 – 20,371,069 |
| Length | 98 |
| Max. P | 0.998927 |

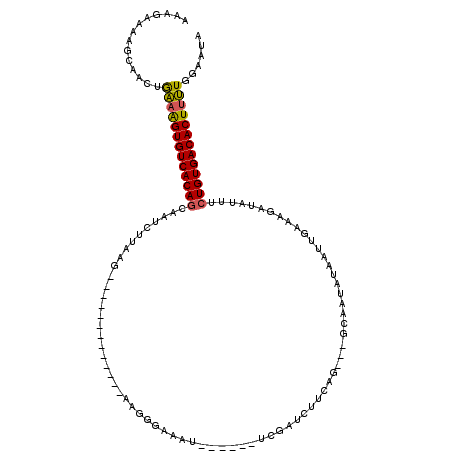
| Location | 20,370,971 – 20,371,069 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.54 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.08 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20370971 98 + 23771897 UAUUCCAAAAGUGUCACAGAAAUAUCUUUCAAUUACAUUGC---UCGAAGAUCGA------AUUUCCUUU-------------CUUAAGAUUGCUGUGACACUUUCAUUUGCCUUUCUUU .......((((((((((((....(((((.............---(((.....)))------.........-------------...)))))..))))))))))))............... ( -22.41) >DroSec_CAF1 5000 98 + 1 UAUUCCAAAAGUGUCACAGAAAUAUUUUUAAAUUAUAUUGC---CUGAGGAUCGA------AUUUCCCAU-------------CUUAAGAUUGCUGUGACACUUUCAGUUGCUUUUCUUU .......((((((((((((.(((((.........)))))..---((.(((((.(.------.....).))-------------))).))....))))))))))))............... ( -20.20) >DroSim_CAF1 5328 98 + 1 UAUUCCAAAAGUGUCACAGAAAUAUUUUUCAAUUAUAUUGC---CCGAAGAUCGA------AUUUCCCAU-------------CUUAAGAUUGCUGUGACACUUUCACUUGCUUUUCUUU .......((((((((((((.(((((.........)))))..---...(((((.(.------.....).))-------------))).......))))))))))))............... ( -19.30) >DroEre_CAF1 4866 85 + 1 UAUUCAAAAAGUGUCACAGAAGUAUCUUUCAAUUAUAUUG----------------------CAACAUUU-------------CUUAAGAUUACUGUGACACUUUUAGUUAGCUUUCUUU ......(((((((((((((....(((((............----------------------........-------------...)))))..))))))))))))).............. ( -19.60) >DroAna_CAF1 2502 120 + 1 CAUUUUAGGAGUGUCACAAAAAUCACUUUAAAAUAUGCUUUUUUGUAAAGAUUUAAAACUAACAAUACUGUAAAAAGAGACUCGUUGAUUUUUAUGUGACACCUCUCGCAAUAAUUCCUU ......((..(((((((((((((((.((((((...(((......)))....))))))....(((....)))..............)))))))..))))))))..)).............. ( -19.80) >consensus UAUUCCAAAAGUGUCACAGAAAUAUCUUUCAAUUAUAUUGC___CCGAAGAUCGA______AUUUCCCUU_____________CUUAAGAUUGCUGUGACACUUUCAGUUGCCUUUCUUU .......((((((((((((....(((((..........................................................)))))..))))))))))))............... (-15.40 = -15.08 + -0.32)

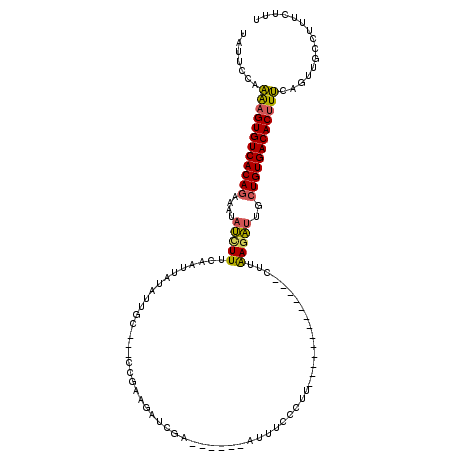
| Location | 20,370,971 – 20,371,069 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.54 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.16 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20370971 98 - 23771897 AAAGAAAGGCAAAUGAAAGUGUCACAGCAAUCUUAAG-------------AAAGGAAAU------UCGAUCUUCGA---GCAAUGUAAUUGAAAGAUAUUUCUGUGACACUUUUGGAAUA ..............(((((((((((((..(((((...-------------........(------(((.....)))---)((((...)))).)))))....)))))))))))))...... ( -23.10) >DroSec_CAF1 5000 98 - 1 AAAGAAAAGCAACUGAAAGUGUCACAGCAAUCUUAAG-------------AUGGGAAAU------UCGAUCCUCAG---GCAAUAUAAUUUAAAAAUAUUUCUGUGACACUUUUGGAAUA ............(..((((((((((((..(((....)-------------))((((...------....))))...---......................))))))))))))..).... ( -24.40) >DroSim_CAF1 5328 98 - 1 AAAGAAAAGCAAGUGAAAGUGUCACAGCAAUCUUAAG-------------AUGGGAAAU------UCGAUCUUCGG---GCAAUAUAAUUGAAAAAUAUUUCUGUGACACUUUUGGAAUA ..............(((((((((((((...(((.(((-------------((.((....------)).))))).))---).......(((....)))....)))))))))))))...... ( -24.60) >DroEre_CAF1 4866 85 - 1 AAAGAAAGCUAACUAAAAGUGUCACAGUAAUCUUAAG-------------AAAUGUUG----------------------CAAUAUAAUUGAAAGAUACUUCUGUGACACUUUUUGAAUA ............(.(((((((((((((..(((((..(-------------(.((((..----------------------..))))..))..)))))....))))))))))))).).... ( -20.10) >DroAna_CAF1 2502 120 - 1 AAGGAAUUAUUGCGAGAGGUGUCACAUAAAAAUCAACGAGUCUCUUUUUACAGUAUUGUUAGUUUUAAAUCUUUACAAAAAAGCAUAUUUUAAAGUGAUUUUUGUGACACUCCUAAAAUG .((((.......(....)((((((((..(((((((..((((..((((((...(((..(............)..))).))))))...)))).....)))))))))))))))))))...... ( -22.00) >consensus AAAGAAAAGCAACUGAAAGUGUCACAGCAAUCUUAAG_____________AAGGGAAAU______UCGAUCUUCAG___GCAAUAUAAUUGAAAGAUAUUUCUGUGACACUUUUGGAAUA ..............(((((((((((((..........................................................................)))))))))))))...... (-14.48 = -14.16 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:45 2006