| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,257,247 – 2,257,391 |
| Length | 144 |
| Max. P | 0.781486 |

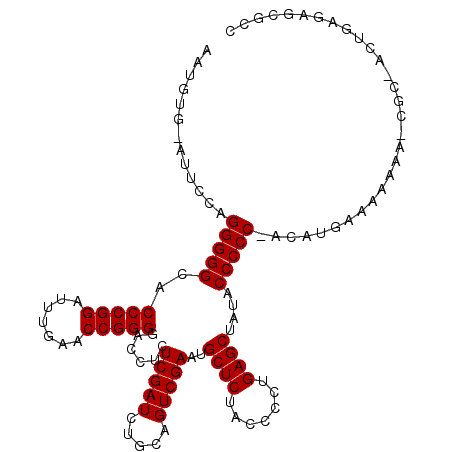
| Location | 2,257,247 – 2,257,358 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.24 |
| Mean single sequence MFE | -35.21 |
| Consensus MFE | -28.53 |
| Energy contribution | -28.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2257247 111 - 23771897 AAUGUA-GUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGAAAAUGG-CGA-ACUCAGAGCGCC ..((((-((((.(((((...(((((.......)))))((.(..(((((.....)))))..).))..)))))))))))))................((-((.-........)))) ( -37.10) >DroVir_CAF1 7285 111 - 1 AAAGUGAAUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCC-AUAUAAAAAACU-UAC-AGUUAGAGCUGU .((((........(((((..(((((.......)))))......(((((.....)))))..((((.......))))....)))))-.........)))-)((-(((....))))) ( -33.33) >DroGri_CAF1 7010 110 - 1 AAUCAA-AUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCC-CAAUGAAAUAUA-CGC-AGUUAGAGCUGC ..((((-((.((.(((....)))))))))))...((((.....(((((.....)))))..((((.......))))......)))-)...........-.((-(((....))))) ( -34.80) >DroMoj_CAF1 6629 111 - 1 AAUAUG-AUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCAAUAUGAAAAAAC-GAG-AUUGGUAGCUAU ......-....((((((...(((((.......)))))((.(..(((((.....)))))..).))..))))))(((((.....(((((.((......)-)..-)))))))))).. ( -33.10) >DroAna_CAF1 7697 111 - 1 AAAGUU-ACCAAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGGUGUUGA-AGC-UCUGGGAGUGCC ......-.......((((..(((((.......)))))..))))(((((.....)))))..((.((.(((..((((((((((........)))))...-)))-)).))))).)). ( -39.40) >DroPer_CAF1 6225 112 - 1 GUUGUG-UUCCAAGGGGGCACCCGGAUUUGAACCGGGGGCCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCAACUUGAAAACAGGCGCCACUGAGA-UGUA ......-...(((((((((.(((((.......))))).)))))(((((.....)))))..((((.......))))...........))))....(((......)))...-.... ( -33.50) >consensus AAUGUG_AUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCC_ACAUGAAAAAAA_CGC_ACUGAGAGCGCC .............(((((..(((((.......)))))......(((((.....)))))..((((.......))))....))))).............................. (-28.53 = -28.53 + 0.00)

| Location | 2,257,285 – 2,257,391 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.15 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2257285 106 + 23771897 CUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUGGAAC-UACAUUUUUUGCCAUAUUUUUUUUUGU-UGGCUGCUC-AAG .(((((((....(((......))).((((....))))(((((.......)))))...)))))))...-..........((((.............-)))).....-... ( -32.22) >DroVir_CAF1 7322 103 + 1 CUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUGGAAUUCACUUUUAU--UUAUUUUUU--AUUUUCCACAUUCUC-AA- ....((((((..(((......))).((((....))))(((((.......))))))))))).(((((...........--.........--...))))).......-..- ( -28.40) >DroEre_CAF1 8132 104 + 1 CUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUGGAAC-CACAUUUUUGGUCAUGUUUU--UUUGC-UGGCUACUC-AAG .....((((((((((...((..(((((((((((((..(((((.......)))))..))...((....-))...)))))))).)))..)--).)))-)..))))))-... ( -33.80) >DroYak_CAF1 7817 105 + 1 CUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUGGAAC-CACAUUUUUGAUCAUAUUUU--UUUGU-GGCCUGCUCUAAG ....(((((((.(((......))).((((((((((..(((((.......)))))..))...((....-))...)))))))).......--.....-...)))))))... ( -32.80) >DroMoj_CAF1 6667 97 + 1 CUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUGGAAU-CAUAUUUUU--UUAU-------AUUACUUCCAUUCCA-GC- ....((((((..(((......))).((((....))))(((((.......)))))))))))(((((((-.........--....-------.........))))))-).- ( -31.25) >DroAna_CAF1 7735 97 + 1 CUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUUUGGU-AACUUUUU----AAUACUUUUUUAUGC-GAACGCC------ ....((((((..(((......))).((((....))))(((((.......))))))))))).......-........----.............((-....)).------ ( -26.50) >consensus CUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCGGUUCAAAUCCGGGUGCCCCCUGGAAC_CACAUUUUU__UCAUAUUUU__UUUGC_UGCCUGCUC_AA_ .(((((((....(((......))).((((....))))(((((.......)))))...)))))))............................................. (-26.82 = -27.15 + 0.33)

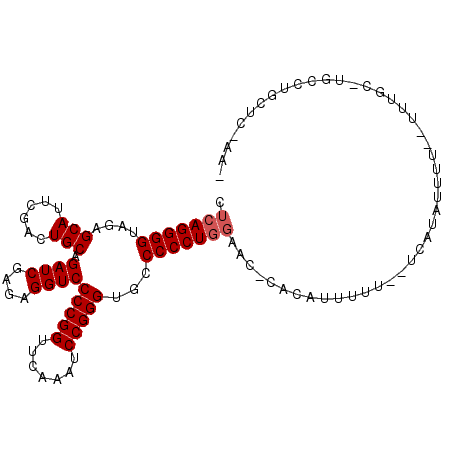
| Location | 2,257,285 – 2,257,391 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -25.87 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2257285 106 - 23771897 CUU-GAGCAGCCA-ACAAAAAAAAAUAUGGCAAAAAAUGUA-GUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAG ...-(((((((((-.............))))......((((-(.((.(((((...(((((.......)))))..))))).)).)))))......))))).......... ( -36.42) >DroVir_CAF1 7322 103 - 1 -UU-GAGAAUGUGGAAAAU--AAAAAAUAA--AUAAAAGUGAAUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAG -..-...............--.........--..............((((((...(((((.......)))))((.(..(((((.....)))))..).))..)))))).. ( -26.70) >DroEre_CAF1 8132 104 - 1 CUU-GAGUAGCCA-GCAAA--AAAACAUGACCAAAAAUGUG-GUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAG ...-.........-.....--.......(((((......))-))).((((((...(((((.......)))))((.(..(((((.....)))))..).))..)))))).. ( -30.30) >DroYak_CAF1 7817 105 - 1 CUUAGAGCAGGCC-ACAAA--AAAAUAUGAUCAAAAAUGUG-GUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAG ..(((((((((((-(((..--................))))-)))....((((..(((((.......)))))..))))(((((.....))))).)))))))........ ( -34.97) >DroMoj_CAF1 6667 97 - 1 -GC-UGGAAUGGAAGUAAU-------AUAA--AAAAAUAUG-AUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAG -..-.....(((((...((-------((..--....)))).-.)))))(((((..(((((.......)))))((.(..(((((.....)))))..).))..)))))... ( -29.20) >DroAna_CAF1 7735 97 - 1 ------GGCGUUC-GCAUAAAAAAGUAUU----AAAAAGUU-ACCAAAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAG ------(((((((-((........(((..----.......)-))....(((((..(((((.......)))))..))))).........).))))))))........... ( -26.90) >consensus _UU_GAGAAGGCA_ACAAA__AAAAUAUGA__AAAAAUGUG_AUUCCAGGGGGCACCCGGAUUUGAACCGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAG ..............................................((((((...(((((.......)))))((.(..(((((.....)))))..).))..)))))).. (-25.87 = -26.03 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:33 2006