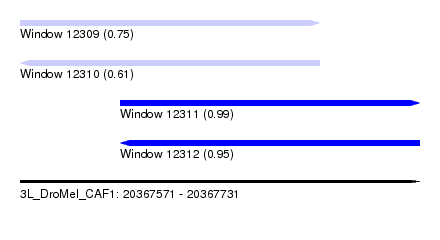
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,367,571 – 20,367,731 |
| Length | 160 |
| Max. P | 0.994995 |

| Location | 20,367,571 – 20,367,691 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -42.91 |
| Consensus MFE | -36.98 |
| Energy contribution | -37.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20367571 120 + 23771897 CCAUUGGCAGGGACCUCUGCGGUUGCAGCAAGGAUAUCUCAUUUUCGUGGCAGCCUUGAUGGCCUUGGUGUUCAUUAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAU .........(((((((((((....))))..)))...))))......(((((((((...(((((((.((((((((((.............))))))).))).)))))))...))))))))) ( -46.62) >DroYak_CAF1 21330 120 + 1 CCAUUGGCAGGGCCCUCUGCGGUUGCAGCAAGGAUAUCUUAUUUUUGUGGCAGCCCUAAUGGCUUUGGUGUUCAUAAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAU ....(((((((((((...((((((((.(((((((((...))))))))).))))))(((..(((...((((((.....)))))).)))....)))...))..))))).......)))))). ( -45.51) >DroAna_CAF1 8841 120 + 1 ACAUUGGCAGGGUCCCCUGAGGUUACAGCAGGGAUAUCUCAUAUUUGUGGCUGCCCUAAUGGCUUUGGUAUUUAUCAAAUACCUACCGGAAUGGACGGCCUGGGCUGUAUUGGCUGCAAU ..((((.((((....)))).(((((((((.(((....)))........(((((.((...(((....((((((.....))))))..)))....)).)))))...))))))...))).)))) ( -36.60) >consensus CCAUUGGCAGGGACCUCUGCGGUUGCAGCAAGGAUAUCUCAUUUUUGUGGCAGCCCUAAUGGCUUUGGUGUUCAUAAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAU ....((((((...((..(((((((...((((((((.....))))))))(((((((.....(((...((((((.....)))))).))).....)).)))))..)))))))..)))))))). (-36.98 = -37.77 + 0.78)

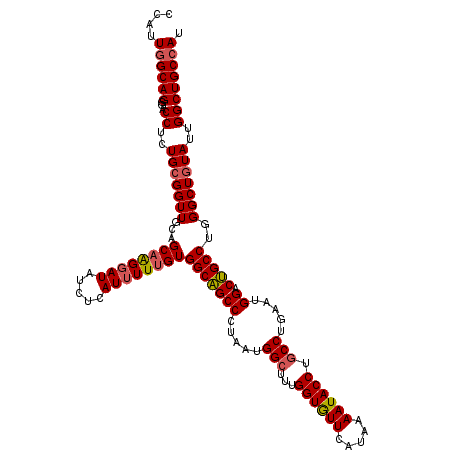
| Location | 20,367,571 – 20,367,691 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -30.99 |
| Energy contribution | -31.67 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20367571 120 - 23771897 AUGGCAGCCAAUACAGCCCAGGCAGUCCAUUCAGGCAGGUAUUUAAUGAACACCAAGGCCAUCAAGGCUGCCACGAAAAUGAGAUAUCCUUGCUGCAACCGCAGAGGUCCCUGCCAAUGG .((((((......((((...(((((((......(((.(((.(((...))).)))...))).....)))))))........(((.....)))))))..(((.....)))..)))))).... ( -34.70) >DroYak_CAF1 21330 120 - 1 AUGGCAGCCAAUACAGCCCAGGCAGUCCAUUCAGGCAGGUAUUUUAUGAACACCAAAGCCAUUAGGGCUGCCACAAAAAUAAGAUAUCCUUGCUGCAACCGCAGAGGGCCCUGCCAAUGG .((((((........((((.((((((((.....(((.(((.(((...))).)))...)))....))))))))....................((((....)))).)))).)))))).... ( -41.30) >DroAna_CAF1 8841 120 - 1 AUUGCAGCCAAUACAGCCCAGGCCGUCCAUUCCGGUAGGUAUUUGAUAAAUACCAAAGCCAUUAGGGCAGCCACAAAUAUGAGAUAUCCCUGCUGUAACCUCAGGGGACCCUGCCAAUGU .(((((((.......((((..((((.......)))).((((((.....))))))..........)))).............((......)))))))))...((((....))))....... ( -31.10) >consensus AUGGCAGCCAAUACAGCCCAGGCAGUCCAUUCAGGCAGGUAUUUAAUGAACACCAAAGCCAUUAGGGCUGCCACAAAAAUGAGAUAUCCUUGCUGCAACCGCAGAGGACCCUGCCAAUGG .((((((.............((((((((.....(((.(((.(((...))).)))...)))....))))))))..............((((..((((....))))))))..)))))).... (-30.99 = -31.67 + 0.67)

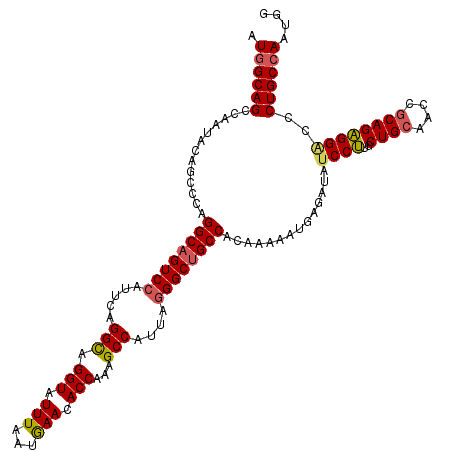
| Location | 20,367,611 – 20,367,731 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -39.94 |
| Consensus MFE | -27.65 |
| Energy contribution | -27.99 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20367611 120 + 23771897 AUUUUCGUGGCAGCCUUGAUGGCCUUGGUGUUCAUUAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUAAGUUUGACACAGCUUGAAGGCCAAGA ......(((((((((...(((((((.((((((((((.............))))))).))).)))))))...))))))))).....(((((..((((((......))))))....))))). ( -44.02) >DroYak_CAF1 21370 118 + 1 AUUUUUGUGGCAGCCCUAAUGGCUUUGGUGUUCAUAAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUGAGCUGGAUGCAGCUUUAAGGGU--GG (((((((.(((((.((....(((...((((((.....)))))).))).....)).))))).(((((((((..(((..(((..(.....)..))))))..).)))))))))))))))--.. ( -43.30) >DroAna_CAF1 8881 108 + 1 AUAUUUGUGGCUGCCCUAAUGGCUUUGGUAUUUAUCAAAUACCUACCGGAAUGGACGGCCUGGGCUGUAUUGGCUGCAAUUUCCAUUUGGGGUAAGCUUU----------UCUCGU--AG .(((..(.(((((((((((.((....((((((.....))))))..))((((..(.(((((...........))))))...))))..))))))).))))..----------.)..))--). ( -32.50) >consensus AUUUUUGUGGCAGCCCUAAUGGCUUUGGUGUUCAUAAAAUACCUGCCUGAAUGGACUGCCUGGGCUGUAUUGGCUGCCAUUUCUAUUUGGGGUAAGCUUGA__CAGCUU_AAGGGU__GG ......(((((((((...((((((..(((((((((...............)))))).)))..))))))...)))))))))((((....))))............................ (-27.65 = -27.99 + 0.34)

| Location | 20,367,611 – 20,367,731 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -21.47 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20367611 120 - 23771897 UCUUGGCCUUCAAGCUGUGUCAAACUUACCCCAAAUAGAAAUGGCAGCCAAUACAGCCCAGGCAGUCCAUUCAGGCAGGUAUUUAAUGAACACCAAGGCCAUCAAGGCUGCCACGAAAAU ..((((((........).)))))..................((((((((......((....))..........(((.(((.(((...))).)))...))).....))))))))....... ( -30.00) >DroYak_CAF1 21370 118 - 1 CC--ACCCUUAAAGCUGCAUCCAGCUCACCCCAAAUAGAAAUGGCAGCCAAUACAGCCCAGGCAGUCCAUUCAGGCAGGUAUUUUAUGAACACCAAAGCCAUUAGGGCUGCCACAAAAAU ..--........(((((....)))))...............((((((((......((....))..........(((.(((.(((...))).)))...))).....))))))))....... ( -30.10) >DroAna_CAF1 8881 108 - 1 CU--ACGAGA----------AAAGCUUACCCCAAAUGGAAAUUGCAGCCAAUACAGCCCAGGCCGUCCAUUCCGGUAGGUAUUUGAUAAAUACCAAAGCCAUUAGGGCAGCCACAAAUAU ..--......----------...(((...(((....((((...((.(((...........))).))...))))(((.((((((.....))))))...)))....))).)))......... ( -23.50) >consensus CC__ACCCUU_AAGCUG__UCAAGCUUACCCCAAAUAGAAAUGGCAGCCAAUACAGCCCAGGCAGUCCAUUCAGGCAGGUAUUUAAUGAACACCAAAGCCAUUAGGGCUGCCACAAAAAU .........................................((((((((......((....))..........(((.(((.(((...))).)))...))).....))))))))....... (-21.47 = -21.70 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:41 2006