
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,355,903 – 20,356,008 |
| Length | 105 |
| Max. P | 0.999624 |

| Location | 20,355,903 – 20,356,008 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Mean single sequence MFE | -18.54 |
| Consensus MFE | -14.06 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20355903 105 + 23771897 CGGCCACCCACUAUUGCCCCCAAGCCGAGAUGUUAUCCAGGAUCUACAGAUCCCAGUUGUCCGCAACCAACUCAACCUACCACAACAAGGACACCAUUAACUACA ((((...................)))).((((...(((.(((((....)))))..((((....)))).....................)))...))))....... ( -18.41) >DroEre_CAF1 25085 105 + 1 CGGCCCCCCACUAUUGCGCCCAAGCCGAGAUGUUAUCCAGGAUCUACAGAUCCUAGUUGUCCGCAACCAACUCAACCUACCACAGUAAGGACACCAUUAACUACA .((..((..(((...((......)).(((.((......((((((....)))))).((((....)))))).)))..........)))..))...)).......... ( -20.10) >DroAna_CAF1 25544 96 + 1 CGCACUCCUACUCCCGCUCCGAAACCCAAUUGCUAUCCAGGAUCAACUGAUCCACGUUGCCCACAAACAAC---------AACUAGAAGGAUUCCAAUCACAACU ............................((((..((((.(((((....)))))..((((........))))---------........))))..))))....... ( -17.10) >consensus CGGCCACCCACUAUUGCCCCCAAGCCGAGAUGUUAUCCAGGAUCUACAGAUCCAAGUUGUCCGCAACCAACUCAACCUACCACAACAAGGACACCAUUAACUACA ............................((((...(((.(((((....)))))..((((........)))).................)))...))))....... (-14.06 = -13.40 + -0.66)

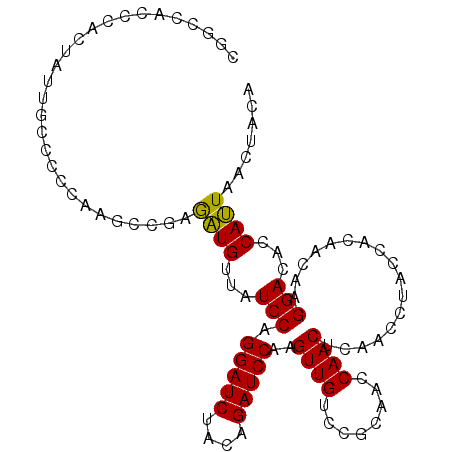
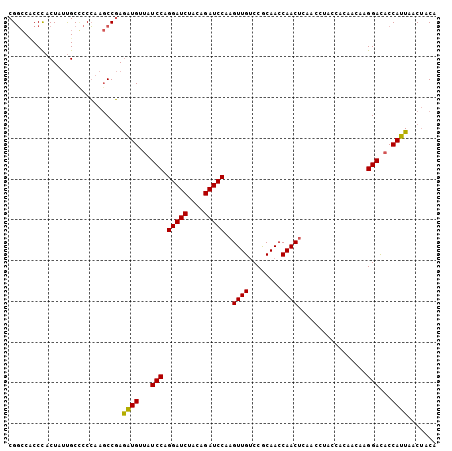
| Location | 20,355,903 – 20,356,008 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Mean single sequence MFE | -32.07 |
| Consensus MFE | -23.52 |
| Energy contribution | -26.20 |
| Covariance contribution | 2.68 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20355903 105 - 23771897 UGUAGUUAAUGGUGUCCUUGUUGUGGUAGGUUGAGUUGGUUGCGGACAACUGGGAUCUGUAGAUCCUGGAUAACAUCUCGGCUUGGGGGCAAUAGUGGGUGGCCG ..........(((..(((((((((..(((((((((.(((((...)))..(..(((((....)))))..)....)).)))))))))...))))))).))..).)). ( -33.30) >DroEre_CAF1 25085 105 - 1 UGUAGUUAAUGGUGUCCUUACUGUGGUAGGUUGAGUUGGUUGCGGACAACUAGGAUCUGUAGAUCCUGGAUAACAUCUCGGCUUGGGCGCAAUAGUGGGGGGCCG ..........(((.((((((((((.((((((((((.(((((...)))..((((((((....))))))))....)).))))))))....)).))))))))))))). ( -37.10) >DroAna_CAF1 25544 96 - 1 AGUUGUGAUUGGAAUCCUUCUAGUU---------GUUGUUUGUGGGCAACGUGGAUCAGUUGAUCCUGGAUAGCAAUUGGGUUUCGGAGCGGGAGUAGGAGUGCG ..((((..(((((((((....((((---------(((((((...(....)..(((((....))))).)))))))))))))))))))).))))............. ( -25.80) >consensus UGUAGUUAAUGGUGUCCUUAUUGUGGUAGGUUGAGUUGGUUGCGGACAACUAGGAUCUGUAGAUCCUGGAUAACAUCUCGGCUUGGGAGCAAUAGUGGGAGGCCG ..........(((.(((((((((((..((((((((.((.(((....)))((((((((....))))))))....)).))))))))...)))))))..)))).))). (-23.52 = -26.20 + 2.68)

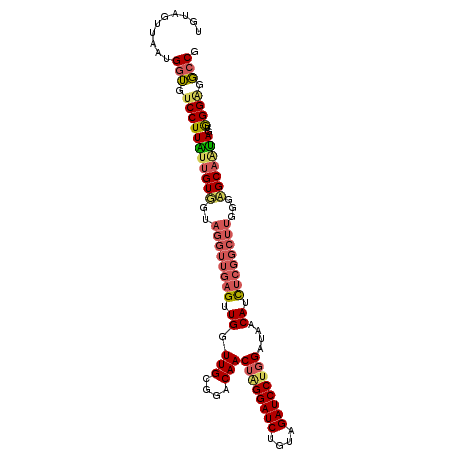
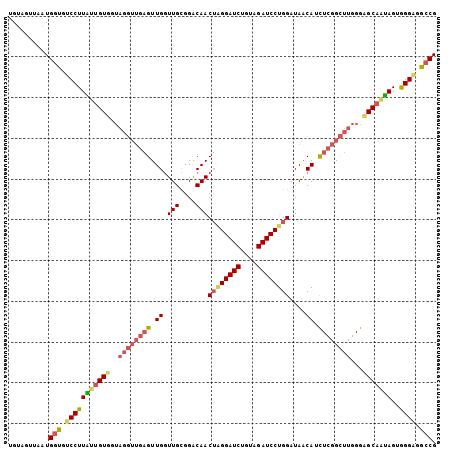
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:38 2006