
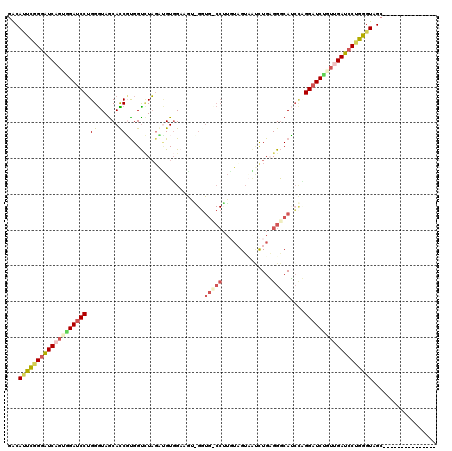
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,354,062 – 20,354,367 |
| Length | 305 |
| Max. P | 0.999258 |

| Location | 20,354,062 – 20,354,167 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.00 |
| Mean single sequence MFE | -40.03 |
| Consensus MFE | -22.00 |
| Energy contribution | -24.44 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
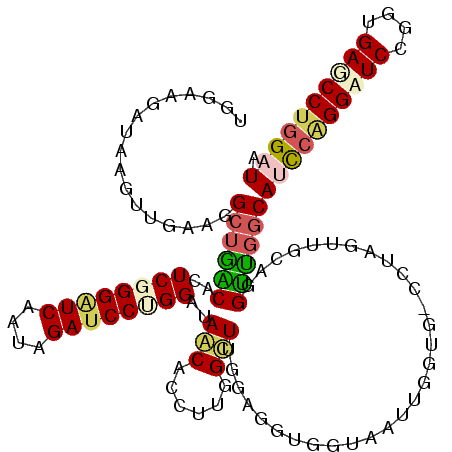
>3L_DroMel_CAF1 20354062 105 - 23771897 GACAUUCGGGGUCAGUGGAUCCUGGGUAGCACCGUGGUCUAGACGUGGAAGUGGGUGCCCUUGUUGUAAUCUGAGGGCAUCCCGGAUCUUUUGAUCCUGGGUAGC--------------- ...((((((((((((.((((((.(..(((.((....)))))..)......(.(((((((((((........)))))))))))))))))).))))))))))))...--------------- ( -46.50) >DroEre_CAF1 23244 102 - 1 GACAUUCGGAAUCAGUAGAUCCUGGGUAGCACCGUGGUCUAGCUGUGGAAGUUGUUGGCCGA---GUAAUUUGAGGGCAUGCUGGAUCUUUUGAUCCUGGGUAGC--------------- ...((((((.(((((.((((((..((((((.((.((((((((((.....)))))..))))).---.........)))).)))))))))).))))).))))))...--------------- ( -32.20) >DroWil_CAF1 30231 93 - 1 CGCACUCAGGAUCGUCUGAACCAGGAUAACAUCUUGGUGUUUGAGUGG------------UUUUAGGCUCCUGUGGGCAACGAGGAUCUGUGGAUCCUGGGUAGC--------------- .((((((((((((((((((((((....(((((....)))))....)))------------.)))))))((((((.....)).))))......)))))))))).))--------------- ( -36.90) >DroAna_CAF1 23382 120 - 1 GACAGCUUGGGUCCGUGGAUCCUGGGUAGCAUCUCGGCUUAGAUGUUGUCAAAGGUGUCCUUGUAGUUGGUUGUGGACAGCUAGGAUCCGUAGAUCCUGGGUAGCAACGUGGCCUAGAUG ........(((((..((((((((.(..(((((((......)))))))..)..((.(((((..............))))).))))))))))..)))))(((((.((...)).))))).... ( -44.54) >consensus GACAUUCGGGAUCAGUGGAUCCUGGGUAGCACCGUGGUCUAGAUGUGGAAGU_GGUG_CCUUGUAGUAAUCUGAGGGCAUCCAGGAUCUGUUGAUCCUGGGUAGC_______________ ...(((((((((((((((((((.((......))......................(((((..............)))))....))))))))))))))))))).................. (-22.00 = -24.44 + 2.44)


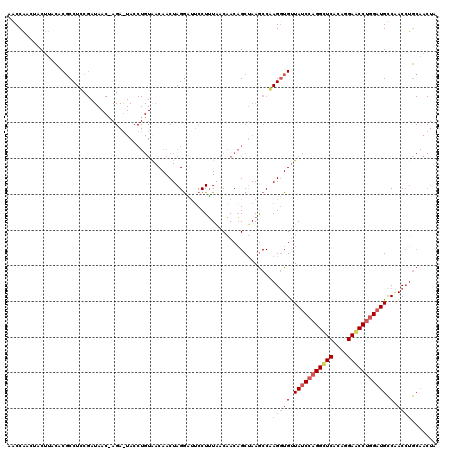
| Location | 20,354,167 – 20,354,287 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.78 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -11.12 |
| Energy contribution | -13.23 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20354167 120 + 23771897 AUCCAACUACUUCCUCGCCUGCGAUAACAAGAAUACCUGUAACAACUAGGAUUCCUUUAACAACAGCUAAGCCCAGGUGUUAUCCAGGUUCACAGGAACCUGGAUGCCAACCUGCAACUA ..............(((....)))......((((.((((.......))))))))...................(((((..(((((((((((....)))))))))))...)))))...... ( -31.00) >DroEre_CAF1 23346 120 + 1 AGCCAGCUACUUACACGCCUCCGAUAACUAGAGUACCUGUGACAACUAGAGUUCCUUUGACAACAGCUAAGCCGAGCUGUUAUCCAGGCUCACAGGAACCUGGAUGCCGACCUGCAACUA ................((...((.((.((((.((.(((((((.(((....)))(((..((.(((((((......))))))).)).))).))))))).)))))).)).))....))..... ( -30.10) >DroWil_CAF1 30324 96 + 1 AACCUAAAACAAAUCCCCCUCC------------------AACAACUAGGCCACCUGUAACUACUGCCAAACCAAGGUGCUAUCCAGGGUCAACAGAUCCCCGCUGUCCACAAA------ ......................------------------........((((((((..................))))).......(((((....)))))..))).........------ ( -12.67) >consensus AACCAACUACUUACACGCCUCCGAUAAC_AGA_UACCUGUAACAACUAGGAUUCCUUUAACAACAGCUAAGCCAAGGUGUUAUCCAGGCUCACAGGAACCUGGAUGCCAACCUGCAACUA ..........................................................................(((((.(((((((((((....))))))))))).).))))....... (-11.12 = -13.23 + 2.11)


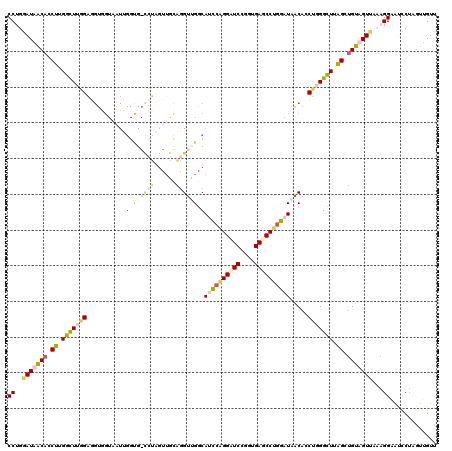
| Location | 20,354,207 – 20,354,327 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.20 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -19.99 |
| Energy contribution | -23.80 |
| Covariance contribution | 3.81 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20354207 120 - 23771897 CCUGGAUAGCAUCUUGGCUUGGAGGUGGUAAUUGGUGGCCUAGUUGCAGGUUGGCAUCCAGGUUCCUGUGAACCUGGAUAACACCUGGGCUUAGCUGUUGUUAAAGGAAUCCUAGUUGUU (((.(((((((.((.(((((..(((((((((((((....))))))))........((((((((((....))))))))))..)))))))))).)).)))))))..)))............. ( -44.80) >DroEre_CAF1 23386 120 - 1 CCCGGAUAGCAUCUUGGCUUGGAGGUGGUAAUUGGUGGCCUAGUUGCAGGUCGGCAUCCAGGUUCCUGUGAGCCUGGAUAACAGCUCGGCUUAGCUGUUGUCAAAGGAACUCUAGUUGUC .((.(((((((.((.((((..(((.((......(.((((((......)))))).)((((((((((....))))))))))..)).))))))).)).)))))))...))............. ( -46.50) >DroWil_CAF1 30346 111 - 1 CCAGGAUAACACCUUGGUUUAGAUGUAGUUCUGGGCG---------UUUGUGGACAGCGGGGAUCUGUUGACCCUGGAUAGCACCUUGGUUUGGCAGUAGUUACAGGUGGCCUAGUUGUU (((((((.(((.((......)).))).)))))))...---------......((((((((((.((....))))))((....(((((.(...((((....)))))))))).))..)))))) ( -30.80) >DroAna_CAF1 23509 105 - 1 CCUGGAUAACACCUAGGCUUUGG---------------UGUUGUUAGGGGCUGGCACUCAGGAUCAGGUGAGCCCGGAUAGCACCUGGGCUUGGCUGUAGUAACAGGGGUCCGUGUGGUU ((((....(((.(((((((..((---------------(((((((..(((((.((.((.......)))).))))).)))))))))..))))))).))).....))))............. ( -44.20) >consensus CCUGGAUAACACCUUGGCUUGGAGGUGGUAAUUGGUG_CCUAGUUGCAGGUUGGCAUCCAGGAUCCGGUGAGCCUGGAUAACACCUGGGCUUAGCUGUAGUUAAAGGAAUCCUAGUUGUU ((..(((((((.((.(((((((...........(.((((((......)))))).)((((((((((....)))))))))).....))))))).)).)))))))...))............. (-19.99 = -23.80 + 3.81)


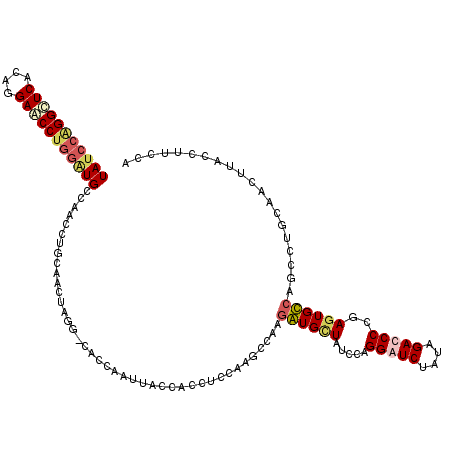
| Location | 20,354,247 – 20,354,367 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.98 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -16.70 |
| Energy contribution | -18.20 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20354247 120 + 23771897 UAUCCAGGUUCACAGGAACCUGGAUGCCAACCUGCAACUAGGCCACCAAUUACCACCUCCAAGCCAAGAUGCUAUCCAGGAUCUCUAGAUCCCGAGUGUCAGCCUUCAACUUACCUUCCG (((((((((((....))))))))))).......((.....(((...................)))..((((((.....(((((....)))))..)))))).))................. ( -32.51) >DroEre_CAF1 23426 120 + 1 UAUCCAGGCUCACAGGAACCUGGAUGCCGACCUGCAACUAGGCCACCAAUUACCACCUCCAAGCCAAGAUGCUAUCCGGGAUCUGUAGACCCCGAGUGUCAGCCUUCAACUUCCCUUCCA .(((((((.((....)).)))))))((.((((((((.((.(((...................))).)).)))....((((.((....)).)))))).))).))................. ( -32.71) >DroWil_CAF1 30386 111 + 1 UAUCCAGGGUCAACAGAUCCCCGCUGUCCACAAA---------CGCCCAGAACUACAUCUAAACCAAGGUGUUAUCCUGGAUCAAGUGAUCCUGAGUGCGUUCCAGCUACAUAUUUACCA ....((((((((...(((((...(((..(.....---------.)..)))....((((((......))))))......)))))...)))))))).((((......)).)).......... ( -28.50) >DroAna_CAF1 23549 105 + 1 UAUCCGGGCUCACCUGAUCCUGAGUGCCAGCCCCUAACAACA---------------CCAAAGCCUAGGUGUUAUCCAGGUUCCAAUGACCCAGAUUGCCAGCCCGCAACGUAUCUUCCA ....((((((.(((((...(((.....)))........((((---------------((........))))))...)))))..((((.......))))..)))))).............. ( -24.40) >consensus UAUCCAGGCUCACAGGAACCUGGAUGCCAACCUGCAACUAGG_CACCAAUUACCACCUCCAAGCCAAGAUGCUAUCCAGGAUCUAUAGACCCCGAGUGCCAGCCUGCAACUUACCUUCCA (((((((((((....))))))))))).........................................((((((.....(((((....)))))..)))))).................... (-16.70 = -18.20 + 1.50)


| Location | 20,354,247 – 20,354,367 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.98 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -23.95 |
| Energy contribution | -25.20 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20354247 120 - 23771897 CGGAAGGUAAGUUGAAGGCUGACACUCGGGAUCUAGAGAUCCUGGAUAGCAUCUUGGCUUGGAGGUGGUAAUUGGUGGCCUAGUUGCAGGUUGGCAUCCAGGUUCCUGUGAACCUGGAUA (....)...........((..((..((((((((....))))))))...(((.((.((((....(((....)))...)))).)).)))..))..))((((((((((....)))))))))). ( -46.60) >DroEre_CAF1 23426 120 - 1 UGGAAGGGAAGUUGAAGGCUGACACUCGGGGUCUACAGAUCCCGGAUAGCAUCUUGGCUUGGAGGUGGUAAUUGGUGGCCUAGUUGCAGGUCGGCAUCCAGGUUCCUGUGAGCCUGGAUA .................((((((..((((((((....))))))))...(((.((.((((....(((....)))...)))).)).)))..))))))((((((((((....)))))))))). ( -49.70) >DroWil_CAF1 30386 111 - 1 UGGUAAAUAUGUAGCUGGAACGCACUCAGGAUCACUUGAUCCAGGAUAACACCUUGGUUUAGAUGUAGUUCUGGGCG---------UUUGUGGACAGCGGGGAUCUGUUGACCCUGGAUA .............((((((((((..(((((....))))).(((((((.(((.((......)).))).))))))))))---------))).....))))((((.((....))))))..... ( -33.70) >DroAna_CAF1 23549 105 - 1 UGGAAGAUACGUUGCGGGCUGGCAAUCUGGGUCAUUGGAACCUGGAUAACACCUAGGCUUUGG---------------UGUUGUUAGGGGCUGGCACUCAGGAUCAGGUGAGCCCGGAUA ..............((((((.((..(((((((..(..(..((((.((((((((........))---------------))))))))))..)..).))))))).....)).)))))).... ( -42.20) >consensus UGGAAGAUAAGUUGAAGGCUGACACUCGGGAUCAAUAGAUCCUGGAUAACACCUUGGCUUGGAGGUGGUAAUUGGUG_CCUAGUUGCAGGUUGGCAUCCAGGAUCCGGUGAGCCUGGAUA .................((((((..((((((((....))))))))..(((......)))..............................))))))((((((((((....)))))))))). (-23.95 = -25.20 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:34 2006