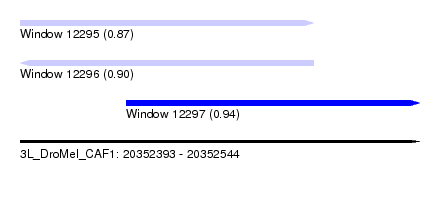
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,352,393 – 20,352,544 |
| Length | 151 |
| Max. P | 0.941464 |

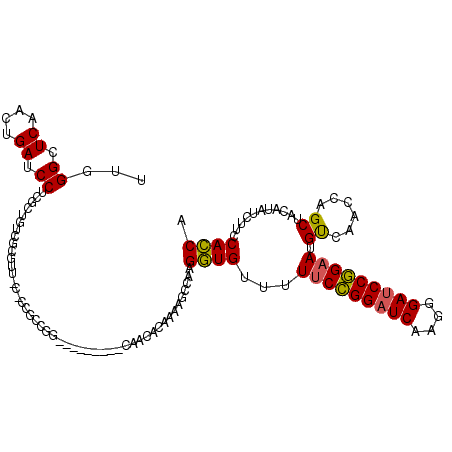
| Location | 20,352,393 – 20,352,504 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.52 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -17.31 |
| Energy contribution | -16.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
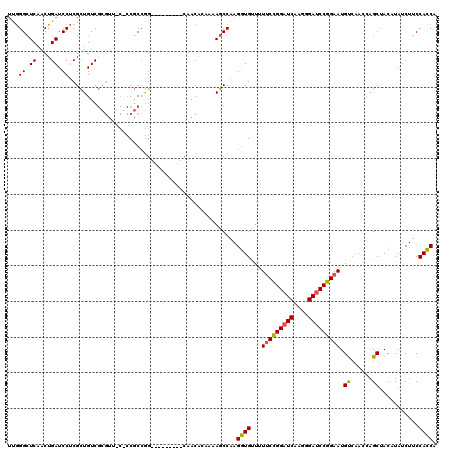
>3L_DroMel_CAF1 20352393 111 + 23771897 UUGGGCUCAACUGAUCCUCGCUGUCGCGUUGCCCCGCCGG---------CAACAAAAAAGCCAAGGUGUUUUUCCGGAUCAAGGGAUCCGGAAUGUCAACCAGCUACCUAUCUUCCACCA ...((((............((....))((((((.....))---------)))).....))))..(((....(((((((((....))))))))).....)))................... ( -35.10) >DroEre_CAF1 21572 111 + 1 UUGGGCUCAACUGAUCCUCGCUGUCGCGUUACGCCGCCGG---------CAACACCAAAGCCAAGGUGUUUUUCCGGAUCAAGGGAUCCGGAAUGUCAACCAGCUACUUAUCUGCCACCA ...(((((....)).....((((..(((......))).((---------(((((((........)))))..(((((((((....)))))))))))))...)))).........))).... ( -35.70) >DroWil_CAF1 28844 105 + 1 CCAGGAUCCACUGAUCCCCGUUGUC---------CUCAAGAAUCAAGAACUACUCUCAAACCAAGAUGCUAUCCUGGUUCAACCGACCCAGAAUGC------GCCACAUAUCUUCCAUCA ...(((((....)))))........---------.....((...((((.....(((.......))).(((((.((((.((....)).)))).))).------))......))))...)). ( -14.90) >consensus UUGGGCUCAACUGAUCCUCGCUGUCGCGUU_C_CCGCCGG_________CAACACAAAAGCCAAGGUGUUUUUCCGGAUCAAGGGAUCCGGAAUGUCAACCAGCUACAUAUCUUCCACCA ...((.((....)).))...............................................((((...(((((((((....))))))))).((......))...........)))). (-17.31 = -16.87 + -0.44)

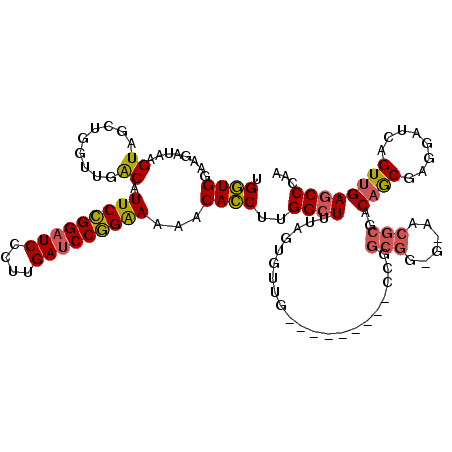
| Location | 20,352,393 – 20,352,504 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.52 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -21.78 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20352393 111 - 23771897 UGGUGGAAGAUAGGUAGCUGGUUGACAUUCCGGAUCCCUUGAUCCGGAAAAACACCUUGGCUUUUUUGUUG---------CCGGCGGGGCAACGCGACAGCGAGGAUCAGUUGAGCCCAA ............((((((((((.....(((((((((....))))))))).....(((((.((.((.(((((---------((.....))))))).)).))))))))))))))..)))... ( -41.20) >DroEre_CAF1 21572 111 - 1 UGGUGGCAGAUAAGUAGCUGGUUGACAUUCCGGAUCCCUUGAUCCGGAAAAACACCUUGGCUUUGGUGUUG---------CCGGCGGCGUAACGCGACAGCGAGGAUCAGUUGAGCCCAA ..(.(((......((.((((((.....(((((((((....))))))))).((((((........)))))))---------))))).))....(((....)))............)))).. ( -42.00) >DroWil_CAF1 28844 105 - 1 UGAUGGAAGAUAUGUGGC------GCAUUCUGGGUCGGUUGAACCAGGAUAGCAUCUUGGUUUGAGAGUAGUUCUUGAUUCUUGAG---------GACAACGGGGAUCAGUGGAUCCUGG .(((...(((.(((....------.))))))..)))..(..(((((((((...)))))))))..)..((.(((((..(...)..))---------))).)).((((((....)))))).. ( -30.70) >consensus UGGUGGAAGAUAAGUAGCUGGUUGACAUUCCGGAUCCCUUGAUCCGGAAAAACACCUUGGCUUUAGUGUUG_________CCGGCGG_G_AACGCGACAGCGAGGAUCAGUUGAGCCCAA .((((........((.........)).(((((((((....)))))))))...))))..((((.....................(((......)))..((((........))))))))... (-21.78 = -22.23 + 0.45)

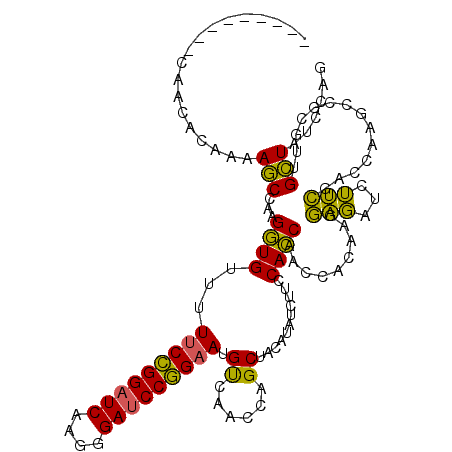
| Location | 20,352,433 – 20,352,544 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.24 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -17.45 |
| Energy contribution | -15.23 |
| Covariance contribution | -2.21 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20352433 111 + 23771897 ---------CAACAAAAAAGCCAAGGUGUUUUUCCGGAUCAAGGGAUCCGGAAUGUCAACCAGCUACCUAUCUUCCACCAACAACGAGGAGAUCUUCGACCAAGCCGCGUUGCUAUCCCG ---------((((.....(((...(((....(((((((((....))))))))).....))).)))...................(((((....)))))..........))))........ ( -28.90) >DroEre_CAF1 21612 111 + 1 ---------CAACACCAAAGCCAAGGUGUUUUUCCGGAUCAAGGGAUCCGGAAUGUCAACCAGCUACUUAUCUGCCACCAACCACAAGGAGGUCUUCGCCCAAGCCGCGUUGCUAUCCAG ---------.((((((........)))))).(((((((((....))))))))).(.(((((.(((........(((.((........)).))).........))).).)))))....... ( -31.03) >DroWil_CAF1 28875 114 + 1 AAUCAAGAACUACUCUCAAACCAAGAUGCUAUCCUGGUUCAACCGACCCAGAAUGC------GCCACAUAUCUUCCAUCAACCACAAGACUACCGGUAACUAAACCAAGAUGUUACCCUG ......................((((((((((.((((.((....)).)))).))).------)).....)))))....................((((((...........))))))... ( -15.70) >consensus _________CAACACAAAAGCCAAGGUGUUUUUCCGGAUCAAGGGAUCCGGAAUGUCAACCAGCUACAUAUCUUCCACCAACCACAAGGAGAUCUUCGACCAAGCCGCGUUGCUAUCCAG ..................(((...((((...(((((((((....))))))))).((......))...........)))).........(((...)))..............)))...... (-17.45 = -15.23 + -2.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:50:27 2006