| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,256,948 – 2,257,071 |
| Length | 123 |
| Max. P | 0.679909 |

| Location | 2,256,948 – 2,257,050 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 86.57 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.36 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
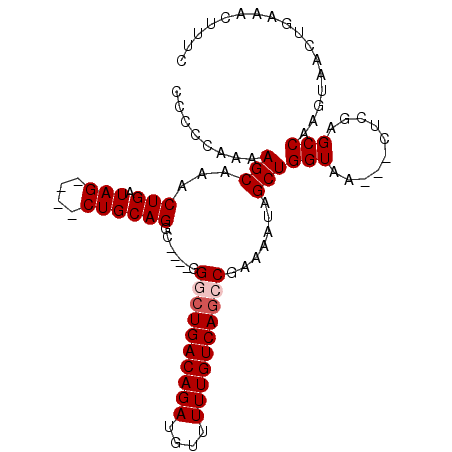
>3L_DroMel_CAF1 2256948 102 - 23771897 CCCCCAAAAGCAAACUGAUAGCUAGCUGCAGACGGACGGACUGACAGAUGUUUUUGUCAUCCGAAAAUAGCUGGUAA---CUCGAGCCAAGAAACUGAAACUUUC ......((((....(((.(((....)))))).(((.((((.(((((((....)))))))))))......(((.(...---..).))).......)))...)))). ( -23.20) >DroSec_CAF1 6812 94 - 1 CACCCAAAAGCAAACUGAUAG----CUGCAGGC----GGACUGACAGAUGUUUUUGUCAGCCGAAAAUAGCUGGUAA---CUCGAGCCAAGUAACUGAAACUUUC .............(((...((----(((....(----((.((((((((....)))))))))))....)))))(((..---.....))).)))............. ( -23.50) >DroSim_CAF1 6891 93 - 1 CCCCCAAAAGCA-ACUGAUAG----CUGCAGGC----GGGCUGACAGAUGUUUUUGUCAGCCGAAAAUAGCUGGUAA---CUCGAGCCAAGUAACUGAAACUUUC ........(((.-.......)----)).(((..----.((((((((((....)))))))))).......((((((..---.....))).)))..)))........ ( -25.10) >DroEre_CAF1 7812 86 - 1 -------AAGCAAACUGAUAG----CUGCAGGC----GGGCUGACAGAUGUUUUUGUCAGCC-AAAAUAGCUGGUAA---CACAAGCCAAGUAACUGAAACUUUC -------.(((.........)----)).(((..----.((((((((((....))))))))))-......((((((..---.....))).)))..)))........ ( -24.30) >DroYak_CAF1 7480 96 - 1 ACUCCGAAAGCAAACUGAUAG----CUGCAGGC----GGGCUGACAGAUGUUUUUGUCAGCC-AAAAUAGCUGGUAACAACACAAGCCAAGUAGAUGAAACUUUC .....(((((...........----((((.(((----.((((((((((....))))))))))-......(.((....)).)....)))..))))......))))) ( -25.73) >consensus CCCCCAAAAGCAAACUGAUAG____CUGCAGGC____GGGCUGACAGAUGUUUUUGUCAGCCGAAAAUAGCUGGUAA___CUCGAGCCAAGUAACUGAAACUUUC ........(((...(((.(((....)))))).......((((((((((....)))))))))).......)))(((..........)))................. (-19.76 = -20.36 + 0.60)

| Location | 2,256,973 – 2,257,071 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -18.10 |
| Energy contribution | -20.10 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
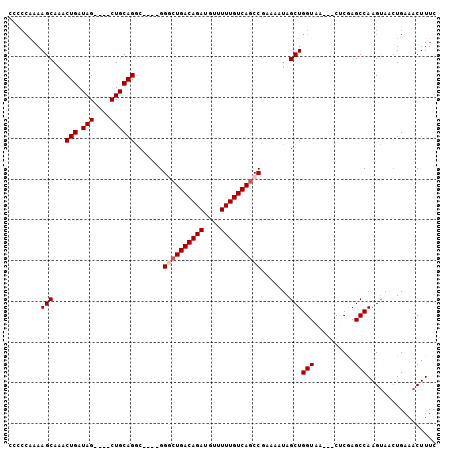
>3L_DroMel_CAF1 2256973 98 + 23771897 --UUACCAGCUAUUUUCGGAUGACAAAAACAUCUGUCAGUCCGUCCGUCUGCAGCUAGCUAUCAGUUUGCUUUUGGGGGAGGGGGCGGGCGAAAGGAGAA --.......((.(((((...(((((........)))))((((((((.(((.(((..(((.........))).))).)))...))))))))))))).)).. ( -30.40) >DroSec_CAF1 6837 90 + 1 --UUACCAGCUAUUUUCGGCUGACAAAAACAUCUGUCAGUCC----GCCUGCAG----CUAUCAGUUUGCUUUUGGGUGGCGGGGCGGGCGAAAGGAGAA --..........(((((((((((((........))))))))(----((((((.(----(((((............))))))...)))))))...))))). ( -34.50) >DroSim_CAF1 6916 89 + 1 --UUACCAGCUAUUUUCGGCUGACAAAAACAUCUGUCAGCCC----GCCUGCAG----CUAUCAGU-UGCUUUUGGGGGCGGGGGCGGGCGAAAGGAGAA --..........(((((((((((((........))))))))(----((((((.(----((..(((.-.....)))..)))....)))))))...))))). ( -36.30) >DroEre_CAF1 7837 78 + 1 --UUACCAGCUAUUUU-GGCUGACAAAAACAUCUGUCAGCCC----GCCUGCAG----CUAUCAGUUUGCUU---------UGG--GGGCGAAAGCAGAA --......(((.....-((((((((........))))))))(----((((.(((----(.........))..---------)).--)))))..))).... ( -28.00) >DroYak_CAF1 7506 88 + 1 UGUUACCAGCUAUUUU-GGCUGACAAAAACAUCUGUCAGCCC----GCCUGCAG----CUAUCAGUUUGCUUUCGGAGU-UUGG--GCGCGAAAGGAGAA .......((((.....-((((((((........)))))))).----......))----)).....(((.((((((..((-....--)).)))))).))). ( -25.00) >consensus __UUACCAGCUAUUUUCGGCUGACAAAAACAUCUGUCAGCCC____GCCUGCAG____CUAUCAGUUUGCUUUUGGGGG_GGGGGCGGGCGAAAGGAGAA .........((.(((((((((((((........)))))))).....((((((......(((..((....))..)))........))))))))))).)).. (-18.10 = -20.10 + 2.00)

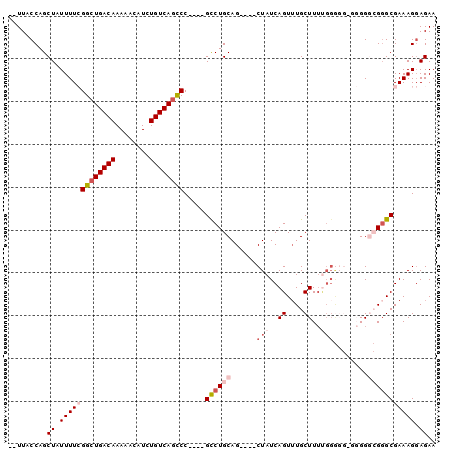
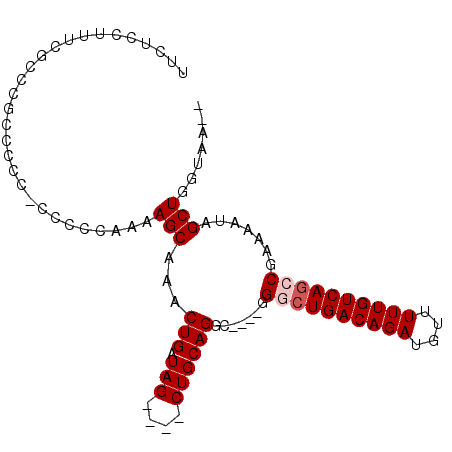
| Location | 2,256,973 – 2,257,071 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
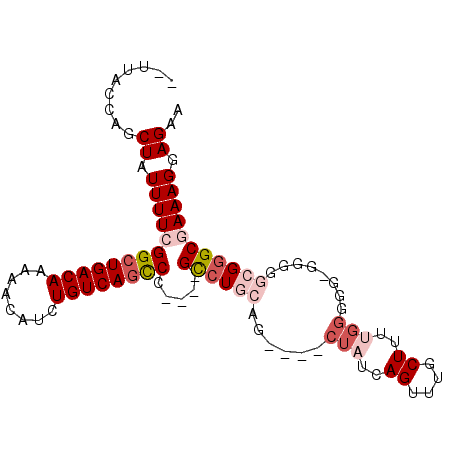
>3L_DroMel_CAF1 2256973 98 - 23771897 UUCUCCUUUCGCCCGCCCCCUCCCCCAAAAGCAAACUGAUAGCUAGCUGCAGACGGACGGACUGACAGAUGUUUUUGUCAUCCGAAAAUAGCUGGUAA-- ..........(((.((...................(((.(((....)))))).....((((.(((((((....)))))))))))......)).)))..-- ( -23.30) >DroSec_CAF1 6837 90 - 1 UUCUCCUUUCGCCCGCCCCGCCACCCAAAAGCAAACUGAUAG----CUGCAGGC----GGACUGACAGAUGUUUUUGUCAGCCGAAAAUAGCUGGUAA-- ..........(((.((.(((((.......(((.........)----))...)))----)).((((((((....)))))))).........)).)))..-- ( -25.20) >DroSim_CAF1 6916 89 - 1 UUCUCCUUUCGCCCGCCCCCGCCCCCAAAAGCA-ACUGAUAG----CUGCAGGC----GGGCUGACAGAUGUUUUUGUCAGCCGAAAAUAGCUGGUAA-- ..........(((.((...((((......(((.-.......)----))...)))----)((((((((((....)))))))))).......)).)))..-- ( -31.00) >DroEre_CAF1 7837 78 - 1 UUCUGCUUUCGCCC--CCA---------AAGCAAACUGAUAG----CUGCAGGC----GGGCUGACAGAUGUUUUUGUCAGCC-AAAAUAGCUGGUAA-- .((.(((..((((.--...---------.(((.........)----))...)))----)((((((((((....))))))))))-.....))).))...-- ( -27.00) >DroYak_CAF1 7506 88 - 1 UUCUCCUUUCGCGC--CCAA-ACUCCGAAAGCAAACUGAUAG----CUGCAGGC----GGGCUGACAGAUGUUUUUGUCAGCC-AAAAUAGCUGGUAACA .....((((((.(.--....-...)))))))........(((----(((.....----.((((((((((....))))))))))-....))))))...... ( -25.90) >consensus UUCUCCUUUCGCCCGCCCCC_CCCCCAAAAGCAAACUGAUAG____CUGCAGGC____GGGCUGACAGAUGUUUUUGUCAGCCGAAAAUAGCUGGUAA__ .............................(((...(((.(((....)))))).......((((((((((....)))))))))).......)))....... (-18.26 = -18.86 + 0.60)

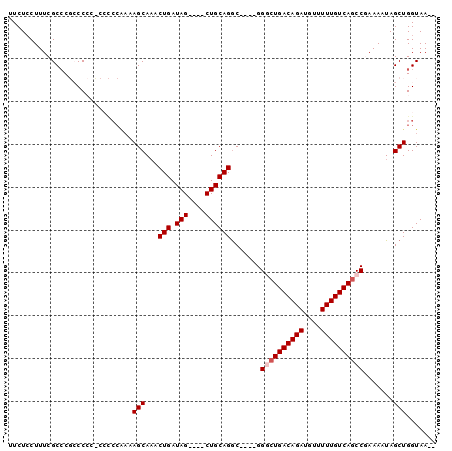
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:30 2006