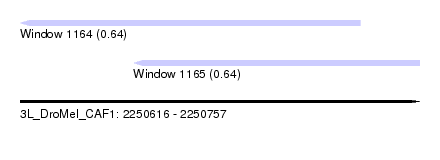
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,250,616 – 2,250,757 |
| Length | 141 |
| Max. P | 0.643538 |

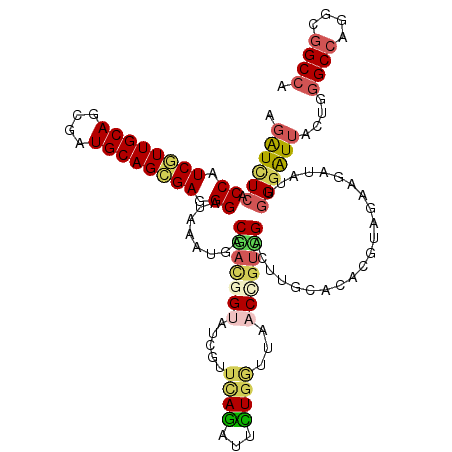
| Location | 2,250,616 – 2,250,736 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -39.91 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2250616 120 - 23771897 AGGUCUCGCCAUCGUUGCACCUGUGCAGCGAGGGGUAGAUGACGAUGGUGUCGUUCAGGUUCUGGUUAACCGUCGACUUGCACACGUAGAAGAUAUUGGGAUUGGUGGGCCAGGCGGCCA .(((((((((.((((((((....))))))))..))).((((.(((((((.....((((...))))...)))))))..((((....))))....))))))))))....((((....)))). ( -41.70) >DroVir_CAF1 1196 120 - 1 AGAUUUCGCCAUCGUUGCAGCGAUGCAGCGAGGGGUAAAUGACCACAGUAUCGUUUAGAUUUUGGUUAACCGUCGACUUGCACACAUAAAAGAUAUUGGGGUUACUGGGCCAGGCGGCGA .....(((((.((((((((....))))))))..(((...(((((.(((((((.((((......((((.......))))........)))).))))))).)))))....)))....))))) ( -39.54) >DroGri_CAF1 1281 120 - 1 AGAUUUCACCAUCAUUGCAGCGAUGCAGUGAAGGAUAAAUGACAACGGUAUCGUUCAGAUUCUGAUUCACCGUUGACUUGCACACGUAGAAGAUGUUGGGAUUACUCGGCCAGGCGGCCA .(((..((((.((((((((....)))))))).))........(((((((.....((((...))))...))))))).....................))..)))....((((....)))). ( -37.00) >DroWil_CAF1 1363 120 - 1 AUAUCUCACCAUCGUUGCAACGAUGCAGCGAUGGAUAUAUAACGAUGGUAUCGUUCAAAUUUUGUUUCACCGGUGCUUUGCAGACAUAGAAAAUGUUUGCAUUGGUGGGCCAUGGUGCCA ........(((((((((((....)))))))))))...........((((((((.........((..(((((((.....(((((((((.....))))))))))))))))..)))))))))) ( -42.90) >DroMoj_CAF1 1342 120 - 1 AGAUCUCCCCGUCGUUGCAGCGAUGCAGCGAUGGAUAAAUGACGACAGAAUCAUUGAGAUUCUGAUUCACCGUCGACUUGCACACGUAGAAUAUGUUGGGAUUACUGGGCCAGGCGGCCA .((((((.(((((((((((....)))))))))))......((((.(((((((.....)))))))((((..(((..........)))..)))).))))))))))....((((....)))). ( -43.50) >DroAna_CAF1 1202 120 - 1 AGAUCUCCCCGUCAUUGCAGCGAUGCAGCGAGGGAUAUAUUACGAGGGAAUCGUUCAGGUUCUGGUUAACUGUCGACUUGCAAACGUAGAAAAUAUUGGGGUUAGAGGGCCAGGCGGCCA ...((((((((....((((....))))(((((.((((....((...((((((.....)))))).))....))))..)))))...............)))))..))).((((....)))). ( -34.80) >consensus AGAUCUCACCAUCGUUGCAGCGAUGCAGCGAGGGAUAAAUGACGACGGUAUCGUUCAGAUUCUGGUUAACCGUCGACUUGCACACGUAGAAGAUAUUGGGAUUACUGGGCCAGGCGGCCA .((((((.((.((((((((....)))))))).))........(((((((.....((((...))))...)))))))......................))))))....((((....)))). (-26.76 = -27.35 + 0.59)

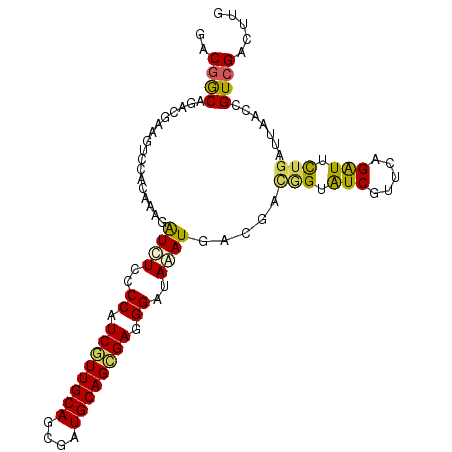
| Location | 2,250,656 – 2,250,757 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.40 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.00 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2250656 101 - 23771897 UGCGGCAGACGUAGUCCACGAAGGUCUCGCCAUCGUUGCACCUGUGCAGCGAGGGGUAGAUGACGAUGGUGUCGUUCAGGUUCUGGUUAACCGUCGACUUG ((((.....))))(((.......((((((((.((((((((....))))))))..))).)).)))((((((.....((((...))))...)))))))))... ( -35.90) >DroVir_CAF1 1236 101 - 1 GACGGCACACGAAAUCCGCAAAGAUUUCGCCAUCGUUGCAGCGAUGCAGCGAGGGGUAAAUGACCACAGUAUCGUUUAGAUUUUGGUUAACCGUCGACUUG (((((.((.(((((((......)))))))((.((((((((....)))))))).))))...((((((....(((.....)))..)))))).)))))...... ( -36.30) >DroGri_CAF1 1321 101 - 1 GACUGCAAACGAAGUUCGAAAAGAUUUCACCAUCAUUGCAGCGAUGCAGUGAAGGAUAAAUGACAACGGUAUCGUUCAGAUUCUGAUUCACCGUUGACUUG ..........((((((......)))))).((.((((((((....)))))))).))........(((((((.....((((...))))...)))))))..... ( -28.40) >DroMoj_CAF1 1382 101 - 1 GACGACAGACGAAAUCCGCAAAGAUCUCCCCGUCGUUGCAGCGAUGCAGCGAUGGAUAAAUGACGACAGAAUCAUUGAGAUUCUGAUUCACCGUCGACUUG ..((((....((.(((......))).)).(((((((((((....))))))))))).....(((...(((((((.....)))))))..)))..))))..... ( -33.70) >DroAna_CAF1 1242 101 - 1 CACGGCACACAUAGUCCAUGAAGAUCUCCCCGUCAUUGCAGCGAUGCAGCGAGGGAUAUAUUACGAGGGAAUCGUUCAGGUUCUGGUUAACUGUCGACUUG ..(((((....(((.((.(((((((..((((.((.(((((....))))).)).))...........))..))).))))))..)))......)))))..... ( -25.70) >DroPer_CAF1 529 101 - 1 AGCGGCAGACAAAGUCCACAAAGGUCUCUCCGUCGUUGCAGCGAUGCAGCGAGGGAUAGAUGACGAUCGUGUCGUUCAGAUUCUGAUUCACCGUCGACUUG ...........(((((.......((((.(((.((((((((....)))))))).))).))))((((...(((....((((...))))..)))))))))))). ( -35.50) >consensus GACGGCAGACGAAGUCCACAAAGAUCUCCCCAUCGUUGCAGCGAUGCAGCGAGGGAUAAAUGACGACGGUAUCGUUCAGAUUCUGAUUAACCGUCGACUUG ..((((.................((((..((.((((((((....)))))))).))..)))).....(((.(((.....))).))).......))))..... (-21.72 = -21.00 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:27 2006