
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,234,467 – 20,234,584 |
| Length | 117 |
| Max. P | 0.905764 |

| Location | 20,234,467 – 20,234,584 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.00 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
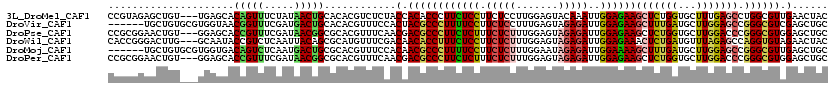
>3L_DroMel_CAF1 20234467 117 + 23771897 CCGUAGAGCUGU---UGAGCACAGUUUCUAUAACUGCACACGUCUCUACCACACCCUUCUCCUUCUCCUUGGAGUACAAAUUGGAGAAGCUCUGGUGCUUUGAGCCUGGCGUUGAACUAC ..((((((((((---.....))))))....((((.((.((.(.(((.....(((((((((((..(((....)))........)))))))....))))....)))).))))))))..)))) ( -30.90) >DroVir_CAF1 3623 114 + 1 ------UGCUGUGCGUGGUAACGGUUUCGAUGACUGCACACGUUUCCACUACGCCCUUUUCCUUCUCCUUUGAGUAGAGAUUGGAGAAGCUUUGAUGCUUGGAGCCGGGCGUCGAGCUGC ------.(((..(((((....(((((.....)))))..))))).....(.((((((((((((.((((.........))))..))))))((((..(...)..)))).)))))).))))... ( -36.90) >DroPse_CAF1 3819 117 + 1 CCGCGGAACUGU---GGAGCACCGUUUCGAUAACGGCGCACGUUUCAACGACGCCCUUCUCUUUCUCUUUGGAGUAGAGAUUGGAGAAGCUCUGGUGCUUGGACCCGGGCGUGGAGCUGC (((((...(((.---((((((((...........((((..(((....))).))))(((((((.(((((.......)))))..)))))))....))))))....))))).)))))...... ( -42.80) >DroWil_CAF1 3824 117 + 1 CACCGGGACUUG---GCAAUACCGUCUCAAUUACAGCGCAUGUUUCGACAACACCUUUCUCCUUCUCUUUGGAGUAGAGAUUGGAGAAACUCUGAUGUUUAGAGCCAGGUGUAGAACUAC ...((..((.((---((..................)).)).))..))...((((((((((((.(((((.......)))))..)))))).((((((...))))))..))))))........ ( -32.57) >DroMoj_CAF1 8252 114 + 1 ------UGCUGUGCGUGGUGACAGUCUCAAUGACUGCGCACGUUUCCACAACGCCCUUUUCCUUCUCUUUGGAAUAGAGAUUGGAAAAGCUUUGAUGCUUGGAGCCGGGCGUUGAGCUGC ------.((((((((((....)((((.....)))))))))).......((((((((((((((.(((((.......)))))..))))))((((..(...)..)))).)))))))))))... ( -49.20) >DroPer_CAF1 3882 117 + 1 CCGCGGAACUGU---GGAGCACCGUUUCGAUAACGGCGCACGUUUCAACGACGCCCUUCUCUUUCUCUUUGGAGUAGAGAUUGGAGAAGCUCUGGUGCUUGGACCCGGGCGUGGAGCUGC (((((...(((.---((((((((...........((((..(((....))).))))(((((((.(((((.......)))))..)))))))....))))))....))))).)))))...... ( -42.80) >consensus CCGCGGAACUGU___GGAGCACCGUUUCGAUAACUGCGCACGUUUCAACAACGCCCUUCUCCUUCUCUUUGGAGUAGAGAUUGGAGAAGCUCUGAUGCUUGGAGCCGGGCGUGGAGCUGC .....................(((((.....)))))............(.((((((((((((.(((((.......)))))..))))))(((((((...))))))).)))))).)...... (-25.27 = -25.00 + -0.27)


| Location | 20,234,467 – 20,234,584 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -24.67 |
| Energy contribution | -23.57 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20234467 117 - 23771897 GUAGUUCAACGCCAGGCUCAAAGCACCAGAGCUUCUCCAAUUUGUACUCCAAGGAGAAGGAGAAGGGUGUGGUAGAGACGUGUGCAGUUAUAGAAACUGUGCUCA---ACAGCUCUACGG ........(((((..((.....)).......(((((((........(((....)))..)))))))))))).((((((..(((..(((((.....)))))..)...---))..)))))).. ( -36.10) >DroVir_CAF1 3623 114 - 1 GCAGCUCGACGCCCGGCUCCAAGCAUCAAAGCUUCUCCAAUCUCUACUCAAAGGAGAAGGAAAAGGGCGUAGUGGAAACGUGUGCAGUCAUCGAAACCGUUACCACGCACAGCA------ ...(((.(((((((...(((..((......))((((((..............)))))))))...)))))).((((.((((.((...(....)...)))))).))))...)))).------ ( -37.34) >DroPse_CAF1 3819 117 - 1 GCAGCUCCACGCCCGGGUCCAAGCACCAGAGCUUCUCCAAUCUCUACUCCAAAGAGAAAGAGAAGGGCGUCGUUGAAACGUGCGCCGUUAUCGAAACGGUGCUCC---ACAGUUCCGCGG ........((((((.((...((((......))))..))..(((((.(((....)))..))))).))))))(((.(((..((((((((((.....))))))))...---))..))).))). ( -40.20) >DroWil_CAF1 3824 117 - 1 GUAGUUCUACACCUGGCUCUAAACAUCAGAGUUUCUCCAAUCUCUACUCCAAAGAGAAGGAGAAAGGUGUUGUCGAAACAUGCGCUGUAAUUGAGACGGUAUUGC---CAAGUCCCGGUG (((.(((.((((((.(((((.......)))))((((((..(((((.......))))).))))))))))))....)))...)))(((((.......)))))...((---(.......))). ( -31.80) >DroMoj_CAF1 8252 114 - 1 GCAGCUCAACGCCCGGCUCCAAGCAUCAAAGCUUUUCCAAUCUCUAUUCCAAAGAGAAGGAAAAGGGCGUUGUGGAAACGUGCGCAGUCAUUGAGACUGUCACCACGCACAGCA------ ...(((((((((((...(((((((......))))......(((((.......))))).)))...)))))))).(....)((((((((((.....)))))......)))))))).------ ( -42.50) >DroPer_CAF1 3882 117 - 1 GCAGCUCCACGCCCGGGUCCAAGCACCAGAGCUUCUCCAAUCUCUACUCCAAAGAGAAAGAGAAGGGCGUCGUUGAAACGUGCGCCGUUAUCGAAACGGUGCUCC---ACAGUUCCGCGG ........((((((.((...((((......))))..))..(((((.(((....)))..))))).))))))(((.(((..((((((((((.....))))))))...---))..))).))). ( -40.20) >consensus GCAGCUCAACGCCCGGCUCCAAGCACCAGAGCUUCUCCAAUCUCUACUCCAAAGAGAAGGAGAAGGGCGUCGUGGAAACGUGCGCAGUUAUCGAAACGGUGCUCA___ACAGCUCCGCGG (((.....((((((........((......))((((((..(((((.......))))).)))))))))))).((....)).)))((((((.....)))))).................... (-24.67 = -23.57 + -1.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:36 2006