
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,229,282 – 20,229,419 |
| Length | 137 |
| Max. P | 0.989149 |

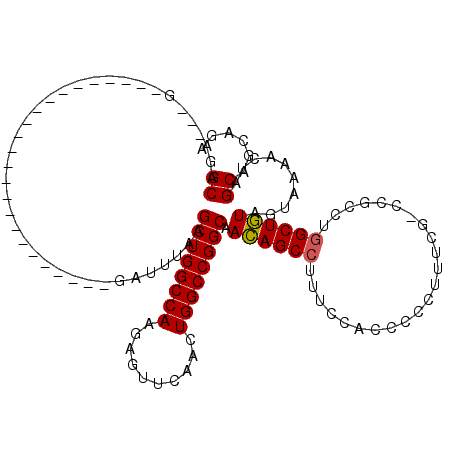
| Location | 20,229,282 – 20,229,379 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20229282 97 - 23771897 GCAGAUGCGG-----------------------GAUUUAAGCUGGCCAAGAGUUCAACUGGCCGGCAACAGCCUUUCCACCCCCUUUCGCCCGCCUUGCUGUAGUAAAACUAGCAGCAGA ......((((-----------------------(....(((..(((((..........)))))(((....)))..........)))...))))).(((((((((.....)).))))))). ( -31.40) >DroSim_CAF1 121 90 - 1 GCAG------------------------------AUUUAAGCUGGCCAAGAGUUCAACUGGCCGGCAACAGCCUUUCCACCCCCUUUCGUCCGCCUGGCUGUAGUAAAACUAGCAGCAGA ((((------------------------------.((((.((((((((..........)))))))).((((((.......................))))))..)))).)).))...... ( -26.50) >DroYak_CAF1 102 116 - 1 GCAGAAUUGAGAUUAAUUCGCUGCGAAUUGUUAGGUUUACGCUGGCCAAGGCUUCAACUGGCCGGCAAUAGCCUUUCCACCCCCUUUCG----CCUGGCUAUAGUAAUACUAGCAGCAGA ...((((((....)))))).((((....((((((..((((((((((((.(.......))))))))).((((((................----...)))))).))))..)))))))))). ( -39.51) >consensus GCAGA___G________________________GAUUUAAGCUGGCCAAGAGUUCAACUGGCCGGCAACAGCCUUUCCACCCCCUUUCG_CCGCCUGGCUGUAGUAAAACUAGCAGCAGA ((......................................((((((((..........)))))))).((((((.......................))))))..........))...... (-23.24 = -23.13 + -0.11)


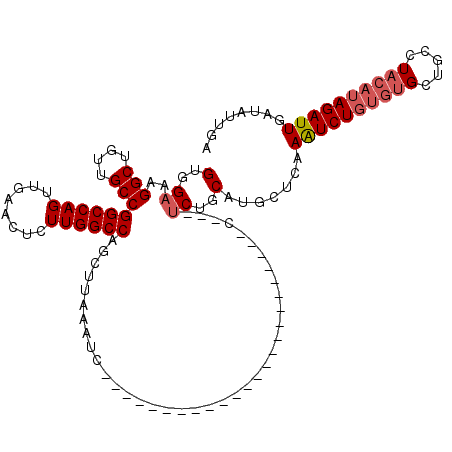
| Location | 20,229,322 – 20,229,419 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.11 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -22.82 |
| Energy contribution | -24.27 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20229322 97 + 23771897 GUGGAAAGGCUGUUGCCGGCCAGUUGAACUCUUGGCCAGCUUAAAUC-----------------------CCGCAUCUGCAUGCUCAAAUCUGUGUGCUGCCUACAUAGAUUGAUAUUGA ((((...(((....)))((((((........))))))..........-----------------------)))).....((...((((.((((((((.....))))))))))))...)). ( -30.30) >DroSim_CAF1 161 90 + 1 GUGGAAAGGCUGUUGCCGGCCAGUUGAACUCUUGGCCAGCUUAAAU------------------------------CUGCAUGCUCAAAUCUGUGUGCUGCCUACAUAGAUUGACAUUGA (..((..(((....)))((((((........))))))........)------------------------------)..)(((.((((.((((((((.....)))))))))))))))... ( -29.90) >DroYak_CAF1 138 116 + 1 GUGGAAAGGCUAUUGCCGGCCAGUUGAAGCCUUGGCCAGCGUAAACCUAACAAUUCGCAGCGAAUUAAUCUCAAUUCUGCUUGCCUAAGUCUGUGUGCUGCCU----AGAUUGAUAUUGG ..((...(((....)))((((((........))))))........))...(((((.((((((((((......))))).((((....))))......)))))..----.)))))....... ( -29.20) >consensus GUGGAAAGGCUGUUGCCGGCCAGUUGAACUCUUGGCCAGCUUAAAUC________________________C___UCUGCAUGCUCAAAUCUGUGUGCUGCCUACAUAGAUUGAUAUUGA (..((..(((....)))((((((........))))))......................................))..).......((((((((((.....))))))))))........ (-22.82 = -24.27 + 1.45)

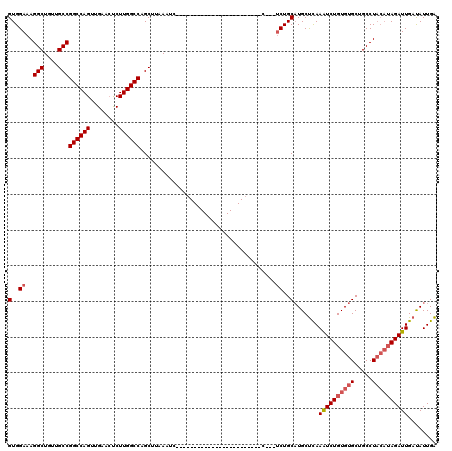
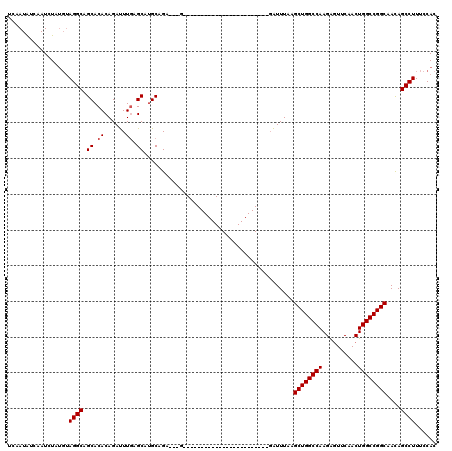
| Location | 20,229,322 – 20,229,419 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.11 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20229322 97 - 23771897 UCAAUAUCAAUCUAUGUAGGCAGCACACAGAUUUGAGCAUGCAGAUGCGG-----------------------GAUUUAAGCUGGCCAAGAGUUCAACUGGCCGGCAACAGCCUUUCCAC .................((((.......((((((..((((....)))).)-----------------------)))))..((((((((..........))))))))....))))...... ( -26.20) >DroSim_CAF1 161 90 - 1 UCAAUGUCAAUCUAUGUAGGCAGCACACAGAUUUGAGCAUGCAG------------------------------AUUUAAGCUGGCCAAGAGUUCAACUGGCCGGCAACAGCCUUUCCAC .................((((.......(((((((......)))------------------------------))))..((((((((..........))))))))....))))...... ( -24.20) >DroYak_CAF1 138 116 - 1 CCAAUAUCAAUCU----AGGCAGCACACAGACUUAGGCAAGCAGAAUUGAGAUUAAUUCGCUGCGAAUUGUUAGGUUUACGCUGGCCAAGGCUUCAACUGGCCGGCAAUAGCCUUUCCAC .((((.((...((----((((........).)))))(((.((.((((((....)))))))))))))))))..(((((...((((((((.(.......)))))))))...)))))...... ( -33.50) >consensus UCAAUAUCAAUCUAUGUAGGCAGCACACAGAUUUGAGCAUGCAGA___G________________________GAUUUAAGCUGGCCAAGAGUUCAACUGGCCGGCAACAGCCUUUCCAC .................((((.((.((......)).))..........................................((((((((..........))))))))....))))...... (-21.30 = -21.63 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:33 2006