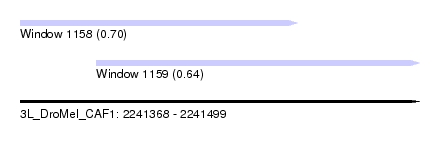
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,241,368 – 2,241,499 |
| Length | 131 |
| Max. P | 0.701189 |

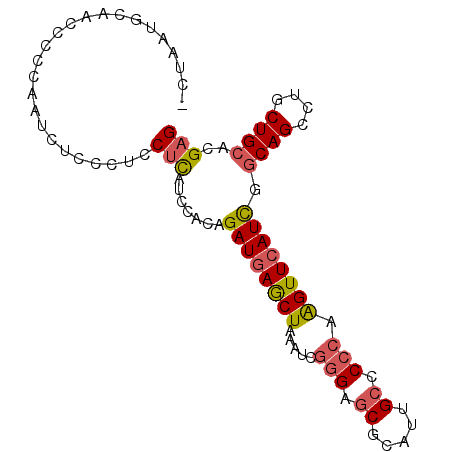
| Location | 2,241,368 – 2,241,459 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2241368 91 + 23771897 -AUAAUGCGACCACCAAUCUCCCGCCUCAUCCACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGUCUGCUGCACGAG -.(((((((.........((((((.((((((....))))))......)))))))))))))............(((((((....)))).))). ( -29.80) >DroGri_CAF1 8376 91 + 1 AAGCAUUCAAA-GAACGUGUUAUUUCACAAUCGCAGAUCAACUGAAUCGCGAACGAAUCGCACUGAGGUUCAUUGACAGCCUGCUGCACGAG ..(((......-....(((......)))....((((.((((.((((((((((.....)))).....))))))))))....)))))))..... ( -20.40) >DroSec_CAF1 2253 91 + 1 -CUAAUGCGACCUCCAAUCUCCCUCCUCAUCCACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGCCUGCUGCACGAG -.(((((((..((((..........((((((....))))))........)))))))))))............(((((((....)))).))). ( -27.77) >DroSim_CAF1 4504 91 + 1 -CUAAUGCGACCUCCAAUCUCCCUCCUCAUCCACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGCCUGCUGCACGAG -.(((((((..((((..........((((((....))))))........)))))))))))............(((((((....)))).))). ( -27.77) >DroEre_CAF1 4997 91 + 1 -CUAAUGCAACUCCCCAUCUCCCUCCUCGUCCGCAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGCCUGCUGCACGAG -.((((((..(((((..........((((((....)))))).......))))).))))))............(((((((....)))).))). ( -27.23) >DroYak_CAF1 5155 91 + 1 -UUAAUGCAGCUCCCCAUCUCCCUUCUUGACCACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGUCUGCUGCACGAG -.((((((.((((((.(((((...(((.......))).)))....)).))))))))))))............(((((((....)))).))). ( -27.30) >consensus _CUAAUGCAACCCCCAAUCUCCCUCCUCAUCCACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGCCUGCUGCACGAG .........................(((.......((((((((.....(((.((.....)).))).)))))))).((((....))))..))) (-18.42 = -18.87 + 0.45)

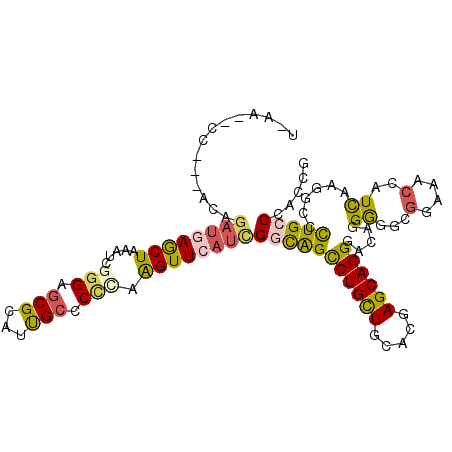
| Location | 2,241,393 – 2,241,499 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -21.49 |
| Energy contribution | -21.47 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2241393 106 + 23771897 UCAU--CC---ACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGUCUGCUGCACGAGCAGGGACAGGGGGCGGAUACCAUCAAGGCCCUGCCCACCG ....--..---...((((((((.....(((.((.....)))))..))))))))....((((.((((....)))))))).((((((((...((.....))..)))))).)). ( -40.60) >DroVir_CAF1 8804 103 + 1 --UAUUUU---UCAGAUCAACUAAAUCGCGAACGAAUCGCGCUAAAGUUCAUUGGCAGCCUGCUUCACGAGCAGGGCGAGGGCAAGGA---GAUUAAGACACUGCCAAUUG --......---.......((((....(((((.....)))))....)))).(((((((((((((((...)))))))((....)).....---..........)))))))).. ( -29.30) >DroEre_CAF1 5022 106 + 1 UCGU--CC---GCAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGCCUGCUGCACGAGCAGGGACAGGGGGCGGAAACUAUCAAGGCCCUGCCCACCG ..((--((---((.((((((((.....(((.((.....)))))..)))))))).))...(((((.....))))))))).((((((((...((....))...)))))).)). ( -46.50) >DroYak_CAF1 5180 106 + 1 UUGA--CC---ACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGUCUGCUGCACGAGCAGGGACAGGGGGCGGAAACCAUCAAGGCCCUGCCCACCG .((.--((---...((((((((.....(((.((.....)))))..))))))))))))((((.((((....)))))))).((((((((...((.....))..)))))).)). ( -41.00) >DroAna_CAF1 7937 109 + 1 AAAC--CCUAAAUAGAUGAGCUAAAUCGGGAGCGCCUCGCGCCGAAGUUCAUCGAUGGCCUGUUGCAUGAGCAGGGUCAGGAAGCGGACACCAUCAAGGCCUUGCCCACGG ....--((......((((((((...((((..(((...))).))))))))))))(((((((((((.....))))))(((.(....).))).)))))..(((...)))...)) ( -38.00) >DroPer_CAF1 2 93 + 1 ------------------AACUAAAUCGGGAGCGCAUUGCCCUUAGGUUCAUUGACAGCCUGCUGCACGAGCAGGGGCAGGAGGCGGGCACAAUCAAAACGCUGCCAAUUG ------------------.........((.((((...(((((....(((....))).((((.((((.(.....)..)))).))))))))).........)))).))..... ( -31.90) >consensus U_AA__CC___ACAGAUGAGCUAAAUCGGGAGCGCAUUGCCCCCAAGUUCAUCGGCAGCCUGCUGCACGAGCAGGGACAGGGGGCGGAAACCAUCAAGGCCCUGCCCACCG ..............((((((((.....(((.(((...))).))).))))))))(((((((((((.....)))))).....((...(....)..))......)))))..... (-21.49 = -21.47 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:18 2006