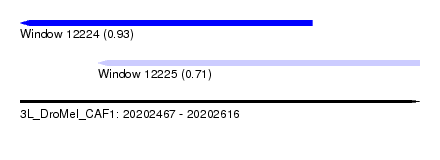
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,202,467 – 20,202,616 |
| Length | 149 |
| Max. P | 0.933205 |

| Location | 20,202,467 – 20,202,576 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.28 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
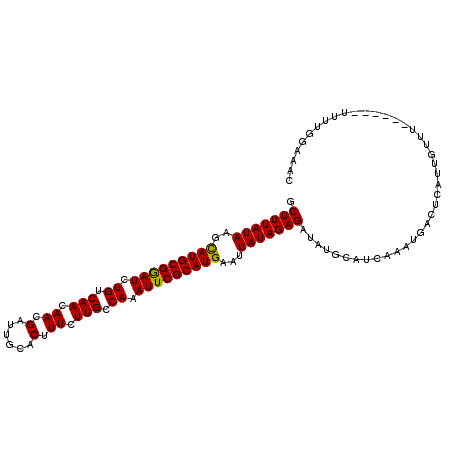
>3L_DroMel_CAF1 20202467 109 - 23771897 GCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGCCAAAUUCGCAUGAAAUAUAGAGAAAUGCAUCAAAUGACUCAUUGUUUUUUUUUUUUUGGAAAC .(((((((..(((((((((.((.(((.((.(......).)).))).)).)))))))))...))))))).((((..((....))..))))((((((........)))))) ( -21.40) >DroSec_CAF1 16368 103 - 1 GCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGACAAAUUCGCAUGAAUUAUAGAGAUAUGCAUCAAAUGACUCAUUGUUU------UUUUGGAAAC .((((((((.(((((((((.((((((.((.(......).)).)))))).)))))))))..)))))))).......(((((.(((.....))).------.))))).... ( -27.40) >DroSim_CAF1 11325 102 - 1 GCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGACAAAUUCGCAUGAAUUAUAGAGAUAUGCAUCAAAUGACUCAUUGUUU-------UUUGGAAAC .((((((((.(((((((((.((((((.((.(......).)).)))))).)))))))))..)))))))).......(((((.(((.....))).-------))))).... ( -28.40) >DroEre_CAF1 11628 102 - 1 GCUUUAUAAGUAUGCGAAUCUGUCAAAAAUGAUUGUACUUUCUUGCCAAAUUCGCAUGAAUUAUAGAGAUAUGCCUUAAAUGAAUCAUUGUU-------UUUUAAAAGC .((((((((.(((((((((.((.(((.((..........)).))).)).)))))))))..))))))))........................-------.......... ( -17.90) >DroYak_CAF1 24970 103 - 1 GCUUUAUAAGCAUGCGGAUCUGUCAAAAACGAUUGUACUUUCUUGCCAAAUUCGCAUGAAUUAUAGAGAUAUGCAUUAAAAGAAUCUUUGUUG------UUUUAAAAAC .((((((((.(((((((((.((.(((.((.(......).)).))).)).)))))))))..)))))))).......((((((.(((....))).------)))))).... ( -21.80) >consensus GCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGCCAAAUUCGCAUGAAUUAUAGAGAUAUGCAUCAAAUGACUCAUUGUUU______UUUUGGAAAC .(((((((..(((((((((.((.(((.((.(......).)).))).)).)))))))))...)))))))......................................... (-18.96 = -18.64 + -0.32)


| Location | 20,202,496 – 20,202,616 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.73 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20202496 120 - 23771897 CAAGUUCAUUUUGACAAAAGAUGCAUUUAUUAUUUAAUUGGCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGCCAAAUUCGCAUGAAAUAUAGAGAAAUGCAUCAAA ((((.....))))......(((((((((.(((......)))(((((((..(((((((((.((.(((.((.(......).)).))).)).)))))))))...))))))))))))))))... ( -28.50) >DroSec_CAF1 16391 118 - 1 CAAGUUGACUUUGUCAUAAGAUACAUUUAUU--UUUAUAGGCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGACAAAUUCGCAUGAAUUAUAGAGAUAUGCAUCAAA ....((((...((((....))))........--..((((..((((((((.(((((((((.((((((.((.(......).)).)))))).)))))))))..)))))))).))))..)))). ( -27.80) >DroSim_CAF1 11347 118 - 1 CAAGUUGACUUUGUCAUAAGAUACAUUUAUU--UUUAUAGGCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGACAAAUUCGCAUGAAUUAUAGAGAUAUGCAUCAAA ....((((...((((....))))........--..((((..((((((((.(((((((((.((((((.((.(......).)).)))))).)))))))))..)))))))).))))..)))). ( -27.80) >DroEre_CAF1 11650 118 - 1 CAAGUUGACUUUGACAACAGAAACAUUUAUU--UUUACUGGCUUUAUAAGUAUGCGAAUCUGUCAAAAAUGAUUGUACUUUCUUGCCAAAUUCGCAUGAAUUAUAGAGAUAUGCCUUAAA ...((((.......)))).............--........((((((((.(((((((((.((.(((.((..........)).))).)).)))))))))..))))))))............ ( -20.20) >DroYak_CAF1 24993 118 - 1 CAAGGUGACUUGGGCAACAAAAACAUUUAUU--UUUACUGGCUUUAUAAGCAUGCGGAUCUGUCAAAAACGAUUGUACUUUCUUGCCAAAUUCGCAUGAAUUAUAGAGAUAUGCAUUAAA ((((....)))).(((..(((((......))--))).....((((((((.(((((((((.((.(((.((.(......).)).))).)).)))))))))..))))))))...)))...... ( -27.40) >consensus CAAGUUGACUUUGACAAAAGAUACAUUUAUU__UUUAUUGGCUUUAUAAGCAUGCGGAUCUGUCAACAACGAUUGCACUUUCUUGCCAAAUUCGCAUGAAUUAUAGAGAUAUGCAUCAAA .........................................(((((((..(((((((((.((.(((.((.(......).)).))).)).)))))))))...)))))))............ (-18.96 = -18.64 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:16 2006