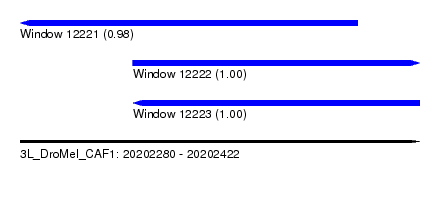
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,202,280 – 20,202,422 |
| Length | 142 |
| Max. P | 0.999920 |

| Location | 20,202,280 – 20,202,400 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -14.54 |
| Energy contribution | -17.66 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20202280 120 - 23771897 AAGUUUCACUAGGAAAAGUAAUUAAAUUACUUUUCUUAGGGAAACUAGACAUACUACUCGCAGUGCCGAAAAUAUUUAUCCUACUUUCCAGAGUGUAAAGUUCUUUUUUGCUGAGAUCAU .((((((.((((((((((((((...)))))))))))))).))))))..........(((((((....(((....(((((...((((....))))))))).)))....)))).)))..... ( -31.40) >DroSec_CAF1 16188 120 - 1 AAGAUUCGCUAGGAAAAGUACAUAGAUUUCUUUUCUUAGGGAAUCUAGACAUACUACUCGCAGUGCCGAAAAUAUUGCUCCUACUCUUCAGAGUGUAGAGUUCUUUUUUUCUGAGAUCGU .((((((.((((((((((...........)))))))))).))))))..........(((((...)).(((((....((((.(((((....)))))..)))).....))))).)))..... ( -32.50) >DroSim_CAF1 11145 120 - 1 AAUUUUCGCUAGGAAAAGUACAUAGAUUUCUUUUCUUAGGGAAUCUAGACAUACUACUCGCAGUGCCGAAAAUAUUGCUCCUACUCUUCAGAGUGUAGAGUUCUUUUUUCCUGAGAUCGU .....((..((((((((((...((((((((((.....)))))))))).)).....((((((((((.......))))))...(((((....)))))..))))....)))))))).)).... ( -31.40) >DroEre_CAF1 11463 99 - 1 UAGUUUCCCUAGGAAAAUUAGUUAAAUUACUUGUCUUAG---------------------CAUUGCCGAAAAGAGUUAUGCUACUUUCCAGAGAGUAUAGUUCUUUUUUGCCGAGAUCGU ((((((..((((.....))))..))))))...((((..(---------------------((.....(((((((..((((((.(((....)))))))))..))))))))))..))))... ( -24.50) >DroYak_CAF1 24784 120 - 1 UAGUUUCCCUAGGCAAAGUAGUUAAGUUACUCGUCUUAGGGAAACUAUUAGUAUUACAUGCAUGGCUGAAAAUUUUUGUCGUACUUUCCAAAGAGUAAAGUUCUUUUUUGCUGAGAUCGU (((((((((((((...(((((.....)))))...)))))))))))))((((((......(((((((...........)))))((((......))))...)).......))))))...... ( -29.82) >consensus AAGUUUCCCUAGGAAAAGUACUUAAAUUACUUUUCUUAGGGAAACUAGACAUACUACUCGCAGUGCCGAAAAUAUUUAUCCUACUUUCCAGAGUGUAAAGUUCUUUUUUGCUGAGAUCGU .((((((.(((((((((((((.....))))))))))))).))))))....................(((.............(((((.(.....).)))))((((.......))))))). (-14.54 = -17.66 + 3.12)


| Location | 20,202,320 – 20,202,422 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -10.90 |
| Energy contribution | -14.82 |
| Covariance contribution | 3.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.46 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20202320 102 + 23771897 GAUAAAUAUUUUCGGCACUGCGAGUAGUAUGUCUAGUUUCCCUAAGAAAAGUAAUUUAAUUACUUUUCCUAGUGAAACUUAAUCAUAUUACUCAUAUCCAAA ((((..........((...))((((((((((...((((((.(((.((((((((((...)))))))))).))).))))))....)))))))))).)))).... ( -32.30) >DroSec_CAF1 16228 101 + 1 GAGCAAUAUUUUCGGCACUGCGAGUAGUAUGUCUAGAUUCCCUAAGAAAAGAAAUCUAUGUACUUUUCCUAGCGAAUCUUAGUC-UAUUACUCAUCUCCGAA ..........(((((......((((((((.(.((((((((.(((.((((((...........)))))).))).))))).))).)-))))))))....))))) ( -27.50) >DroSim_CAF1 11185 101 + 1 GAGCAAUAUUUUCGGCACUGCGAGUAGUAUGUCUAGAUUCCCUAAGAAAAGAAAUCUAUGUACUUUUCCUAGCGAAAAUUAGUC-UAUUACUCAUCUCCGAA ..........(((((......((((((((.(.((((.(((.(((.((((((...........)))))).))).)))..)))).)-))))))))....))))) ( -22.40) >DroEre_CAF1 11503 80 + 1 CAUAACUCUUUUCGGCAAUG---------------------CUAAGACAAGUAAUUUAACUAAUUUUCCUAGGGAAACUAAGUC-UAUUACUCUUCUGAGAA .....(((.....(((...)---------------------)).(((..((((((...(((.(.(((((...))))).).))).-.))))))..)))))).. ( -13.30) >DroYak_CAF1 24824 101 + 1 GACAAAAAUUUUCAGCCAUGCAUGUAAUACUAAUAGUUUCCCUAAGACGAGUAACUUAACUACUUUGCCUAGGGAAACUAAGUC-UAUUACUUUUAUCAAGA ((.((((.......((...))..((((((....(((((((((((.(..(((((.......)))))..).)))))))))))....-)))))))))).)).... ( -23.30) >consensus GAGAAAUAUUUUCGGCACUGCGAGUAGUAUGUCUAGAUUCCCUAAGAAAAGUAAUUUAACUACUUUUCCUAGCGAAACUUAGUC_UAUUACUCAUCUCCGAA .....................((((((((.....((((((.(((.((((((((.......)))))))).))).))))))......))))))))......... (-10.90 = -14.82 + 3.92)


| Location | 20,202,320 – 20,202,422 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -13.13 |
| Energy contribution | -17.81 |
| Covariance contribution | 4.68 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.48 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20202320 102 - 23771897 UUUGGAUAUGAGUAAUAUGAUUAAGUUUCACUAGGAAAAGUAAUUAAAUUACUUUUCUUAGGGAAACUAGACAUACUACUCGCAGUGCCGAAAAUAUUUAUC (((((.(.((((((.((((.((.((((((.((((((((((((((...)))))))))))))).)))))).)))))).)))))).)...))))).......... ( -35.20) >DroSec_CAF1 16228 101 - 1 UUCGGAGAUGAGUAAUA-GACUAAGAUUCGCUAGGAAAAGUACAUAGAUUUCUUUUCUUAGGGAAUCUAGACAUACUACUCGCAGUGCCGAAAAUAUUGCUC (((((...((((((.((-..(((.(((((.((((((((((...........)))))))))).))))))))...)).)))))).....))))).......... ( -31.90) >DroSim_CAF1 11185 101 - 1 UUCGGAGAUGAGUAAUA-GACUAAUUUUCGCUAGGAAAAGUACAUAGAUUUCUUUUCUUAGGGAAUCUAGACAUACUACUCGCAGUGCCGAAAAUAUUGCUC (((((...((((((.((-..(((...(((.((((((((((...........)))))))))).)))..)))...)).)))))).....))))).......... ( -27.10) >DroEre_CAF1 11503 80 - 1 UUCUCAGAAGAGUAAUA-GACUUAGUUUCCCUAGGAAAAUUAGUUAAAUUACUUGUCUUAG---------------------CAUUGCCGAAAAGAGUUAUG ..((.(((.(((((((.-((((...(((((...)))))...))))..))))))).))).))---------------------(((.(((.....).)).))) ( -14.80) >DroYak_CAF1 24824 101 - 1 UCUUGAUAAAAGUAAUA-GACUUAGUUUCCCUAGGCAAAGUAGUUAAGUUACUCGUCUUAGGGAAACUAUUAGUAUUACAUGCAUGGCUGAAAAUUUUUGUC ....(((((((((((((-....(((((((((((((...(((((.....)))))...)))))))))))))....))))))..((...)).......))))))) ( -28.80) >consensus UUCGGAGAUGAGUAAUA_GACUAAGUUUCCCUAGGAAAAGUACUUAAAUUACUUUUCUUAGGGAAACUAGACAUACUACUCGCAGUGCCGAAAAUAUUUAUC (((((....(((((.........((((((.(((((((((((((.....))))))))))))).))))))........)))))......))))).......... (-13.13 = -17.81 + 4.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:14 2006