| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,196,356 – 20,196,459 |
| Length | 103 |
| Max. P | 0.807660 |

| Location | 20,196,356 – 20,196,459 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 73.38 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20196356 103 - 23771897 AAAC-UAU---CUUAUUCCGCAAAAUACCUGCUGCCACUCGGUUUUGUCCAGUGGUUCGCCAUCCGUAAGUGGCGGUCGCAAAAUGGACACGUACUGUGAGUUUUAA ((((-(..---........(((.......))).....(.((((..((((((.(((..((((((......))))))..).))...))))))...)))).))))))... ( -27.60) >DroPse_CAF1 1419 94 - 1 AAAG-CCU---UCUUUGUGG--AUGUACCUGUUGCCAUUCGGUUUUAUCCAGAGGCUCGCCGUCUGUGAGCGGUGGUCGCAGAAUGGAUACGUACUGGAA------- ..((-(((---(.....(((--(((.(((...........)))..))))))))))))((((((......)))))).((.(((.(((....))).))))).------- ( -28.00) >DroGri_CAF1 3877 100 - 1 AUUGCUGUAUUCUUAUAU---AUCAUACCUGUUGCCACUCUCCCUUGUCCAGAGCCUCGCCAUCAGUAAGAGGUGGCCGCAGUAUGGAUACGUACUACAA----UAA ((((..((((.....(((---.((((((.(((.((((((((..((.((...(.....)...)).))..))).))))).)))))))))))).))))..)))----).. ( -23.50) >DroEre_CAF1 5120 101 - 1 AAAC-AAU---CUUAUCCAGGA--AUACCUGCUGCCACUCGGUUUUGUCCAGUGGUUCGCCAUCCGUGAGCGGCGGUCGCAAAAUGGACACGUACUGGGAGUUUUAA ((((-...---......((((.--...))))......((((((..((((((.(((..((((...(....).))))..).))...))))))...)))))).))))... ( -30.80) >DroYak_CAF1 18570 101 - 1 AAAC-UAU---CUUAUCCAGCA--AUACCUGCUGCCACUCGGUUUUGUCCAGUGGUUCGCCAUCCGUAAGUGGUGGUCGCAAGAUGGACACGUACUGAGAAUUUUAA ....-...---......(((((--.....)))))...((((((..((((((...((..(((((((....).)))))).))....))))))...))))))........ ( -33.00) >DroPer_CAF1 1485 94 - 1 AAAG-CCU---UCUUUGUGG--AUGUACCUGUUGCCAUUCGGUUUUAUCCAGAGGCUCGCCGUCUGUGAGCGGUGGUCGCAGAAUGGAUACGUACUGGAA------- ..((-(((---(.....(((--(((.(((...........)))..))))))))))))((((((......)))))).((.(((.(((....))).))))).------- ( -28.00) >consensus AAAC_UAU___CUUAUCCAG_AA_AUACCUGCUGCCACUCGGUUUUGUCCAGAGGCUCGCCAUCCGUAAGCGGUGGUCGCAAAAUGGACACGUACUGGAA____UAA .....................................((((((..((((((...(((((((((......)))))))..))....))))))...))))))........ (-20.10 = -20.18 + 0.08)


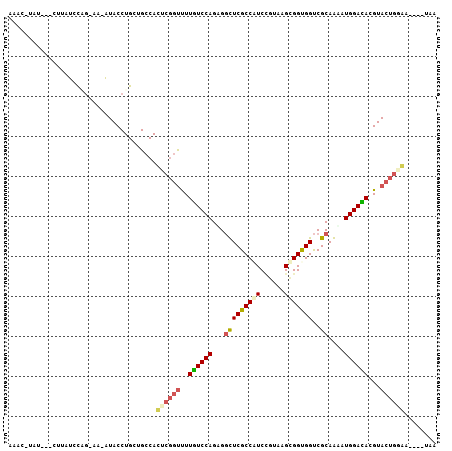
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:11 2006