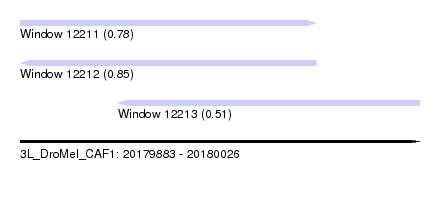
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,179,883 – 20,180,026 |
| Length | 143 |
| Max. P | 0.846233 |

| Location | 20,179,883 – 20,179,989 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -34.09 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
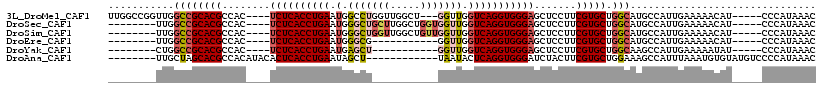
>3L_DroMel_CAF1 20179883 106 + 23771897 GUUUAUGGG-----AUGUUUUUCAAUGGCAUGCCAGCACGAAGGAGCUCCCACCUGACCAACC---AGCCAACCAGGCCAUUCAGGUGAGA----GUGGCGUGCGGCCAACCGGCCAA .....(((.-----(((((.......))))).)))(((((.....((((.(((((((......---.(((.....)))...))))))).))----))..)))))((((....)))).. ( -44.10) >DroSec_CAF1 12330 101 + 1 GUUUAUGGG-----AUGUUUUUCAAUGGCAUGCCAGCACGAAGGAGCUCCCACCUGACCAACCACCAGCCAAGCAGCCCAUUCAGGUGAGA----GUGGCGUGCGGCCAA-------- .....(((.-----(((((.......))))).)))(((((.....((((.(((((((........................))))))).))----))..)))))......-------- ( -31.56) >DroSim_CAF1 13304 101 + 1 GUUUAUGGG-----AUGUUUUUCAAUGGCAUGCCAGCACGAAGGAGCUCCCACCUGACCAACCAACAGCCAACCAGCCCAUUCAGGUGAGA----GUGGCGUGCGGCCAA-------- .....(((.-----(((((.......))))).)))(((((.....((((.(((((((........................))))))).))----))..)))))......-------- ( -31.56) >DroEre_CAF1 12469 90 + 1 GUUUAUGGG-----AUGUUUUUCAAUGGCAUGCCAGCACGAAGGAGCUCCCACCUGACCAACC-----------CGCCCAUUCAGGUGAGA----GUGGCGUGCGGCCAA-------- .....(((.-----(((((.......))))).)))(((((.....((((.(((((((......-----------.......))))))).))----))..)))))......-------- ( -32.22) >DroYak_CAF1 12151 90 + 1 GUUUAUGGG-----AUAUUUUUCAAUGGCUUGCCAGCACGAAGGAGCUCCCACCUGACCAACC-----------AGCUCAUUCAGGUGAGA----GUGGCGUGCGGCCAG-------- .....((..-----(....)..)).(((((.....(((((.....((((.(((((((......-----------.......))))))).))----))..)))))))))).-------- ( -30.92) >DroAna_CAF1 14865 98 + 1 GUUUAUGGGGACAUACACAUUUAAAUGGCUUUCCAGCACGAAGUAGAUCCCACCUGAGUAUUA------------AGCUAUUCAGGUGAGUGUAUGUGGCGUGCUAGCAA-------- ((.((((....)))).)).........(((....((((((..(((.((..((((((((((...------------...)))))))))).)).)))....)))))))))..-------- ( -34.20) >consensus GUUUAUGGG_____AUGUUUUUCAAUGGCAUGCCAGCACGAAGGAGCUCCCACCUGACCAACC___________AGCCCAUUCAGGUGAGA____GUGGCGUGCGGCCAA________ .....(((.................(((....)))(((((.......((.(((((((........................))))))).))........)))))..)))......... (-22.37 = -22.70 + 0.33)


| Location | 20,179,883 – 20,179,989 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -33.59 |
| Consensus MFE | -25.86 |
| Energy contribution | -27.46 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
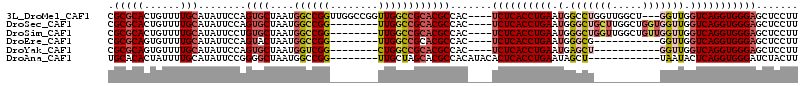
>3L_DroMel_CAF1 20179883 106 - 23771897 UUGGCCGGUUGGCCGCACGCCAC----UCUCACCUGAAUGGCCUGGUUGGCU---GGUUGGUCAGGUGGGAGCUCCUUCGUGCUGGCAUGCCAUUGAAAAACAU-----CCCAUAAAC .(((..(((..((((((((...(----((((((((((.(((((.....))))---).....)))))))))))......))))).)))..))).((....))...-----.)))..... ( -41.60) >DroSec_CAF1 12330 101 - 1 --------UUGGCCGCACGCCAC----UCUCACCUGAAUGGGCUGCUUGGCUGGUGGUUGGUCAGGUGGGAGCUCCUUCGUGCUGGCAUGCCAUUGAAAAACAU-----CCCAUAAAC --------...((((((((...(----((((((((((.(.(((..((.....))..))).))))))))))))......))))).))).................-----......... ( -38.20) >DroSim_CAF1 13304 101 - 1 --------UUGGCCGCACGCCAC----UCUCACCUGAAUGGGCUGGUUGGCUGUUGGUUGGUCAGGUGGGAGCUCCUUCGUGCUGGCAUGCCAUUGAAAAACAU-----CCCAUAAAC --------...((((((((...(----((((((((((.(.((((..(.....)..)))).))))))))))))......))))).))).................-----......... ( -35.00) >DroEre_CAF1 12469 90 - 1 --------UUGGCCGCACGCCAC----UCUCACCUGAAUGGGCG-----------GGUUGGUCAGGUGGGAGCUCCUUCGUGCUGGCAUGCCAUUGAAAAACAU-----CCCAUAAAC --------((((((((.((((.(----..((....))..)))))-----------.).)))))))((((((.....((((...(((....))).)))).....)-----))))).... ( -30.00) >DroYak_CAF1 12151 90 - 1 --------CUGGCCGCACGCCAC----UCUCACCUGAAUGAGCU-----------GGUUGGUCAGGUGGGAGCUCCUUCGUGCUGGCAAGCCAUUGAAAAAUAU-----CCCAUAAAC --------(((((((...(((((----((..........))).)-----------))))))))))((((((.....((((...(((....))).)))).....)-----))))).... ( -30.10) >DroAna_CAF1 14865 98 - 1 --------UUGCUAGCACGCCACAUACACUCACCUGAAUAGCU------------UAAUACUCAGGUGGGAUCUACUUCGUGCUGGAAAGCCAUUUAAAUGUGUAUGUCCCCAUAAAC --------...((((((((.........(((((((((.((...------------...)).)))))))))........)))))))).............((((........))))... ( -26.63) >consensus ________UUGGCCGCACGCCAC____UCUCACCUGAAUGGGCU___________GGUUGGUCAGGUGGGAGCUCCUUCGUGCUGGCAUGCCAUUGAAAAACAU_____CCCAUAAAC ...........((((((((........((((((((((.(.(((((((.....))))))).))))))))))).......))))).)))............................... (-25.86 = -27.46 + 1.60)


| Location | 20,179,918 – 20,180,026 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.93 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -25.92 |
| Energy contribution | -27.77 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20179918 108 - 23771897 CGCGCACUGUUUUGCAUAUUCCAGUGCUAAUGGCCGGUUGGCCGGUUGGCCGCACGCCAC----UCUCACCUGAAUGGCCUGGUUGGCU---GGUUGGUCAGGUGGGAGCUCCUU ...((((((............))))))...((((.(..(((((....)))))..)))))(----((((((((((.(((((.....))))---).....)))))))))))...... ( -46.40) >DroSec_CAF1 12365 103 - 1 CGCGCACUGUUUUGCAUAUUCCAGUGCUAAUGGCCGG--------UUGGCCGCACGCCAC----UCUCACCUGAAUGGGCUGCUUGGCUGGUGGUUGGUCAGGUGGGAGCUCCUU .((((((((............))))))....((((..--------..))))))......(----((((((((((.(.(((..((.....))..))).))))))))))))...... ( -43.60) >DroSim_CAF1 13339 103 - 1 CGCGCACUGUUUUGCAUAUUCCUGUGCUAAUGGCCGG--------UUGGCCGCACGCCAC----UCUCACCUGAAUGGGCUGGUUGGCUGUUGGUUGGUCAGGUGGGAGCUCCUU .(((((......)))........((((....((((..--------..))))))))))..(----((((((((((.(.((((..(.....)..)))).))))))))))))...... ( -38.20) >DroEre_CAF1 12504 92 - 1 CGCGCAGUGUUUUGCAUAUUCCAGUACUAAUGGCCGG--------UUGGCCGCACGCCAC----UCUCACCUGAAUGGGCG-----------GGUUGGUCAGGUGGGAGCUCCUU .((((((....))))................((((..--------..))))))......(----((((((((((.(.((..-----------..)).))))))))))))...... ( -31.20) >DroYak_CAF1 12186 92 - 1 CGCGCAGUGUUUUGCAUAUUCCAGUGCUAAUGGUCGG--------CUGGCCGCACGCCAC----UCUCACCUGAAUGAGCU-----------GGUUGGUCAGGUGGGAGCUCCUU .((((((....))))........((((....((((..--------..))))))))))..(----((((((((((.(.((..-----------..)).))))))))))))...... ( -32.00) >DroAna_CAF1 14905 95 - 1 UGCACACUAUUUUGCAUAUUCCGGGGCUAAUGGCCGG--------UUGCUAGCACGCCACAUACACUCACCUGAAUAGCU------------UAAUACUCAGGUGGGAUCUACUU ((((........))))........(((.....)))((--------(((....)).))).......(((((((((.((...------------...)).)))))))))........ ( -25.10) >consensus CGCGCACUGUUUUGCAUAUUCCAGUGCUAAUGGCCGG________UUGGCCGCACGCCAC____UCUCACCUGAAUGGGCU___________GGUUGGUCAGGUGGGAGCUCCUU .(((((......)))........((((....((((((........)))))))))))).......((((((((((.(.(((((((.....))))))).)))))))))))....... (-25.92 = -27.77 + 1.85)
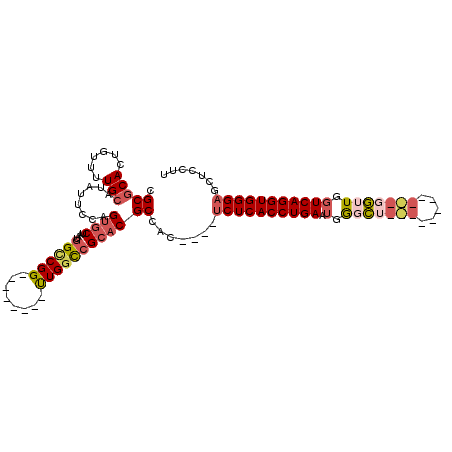
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:49:04 2006