
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,167,419 – 20,167,571 |
| Length | 152 |
| Max. P | 0.998386 |

| Location | 20,167,419 – 20,167,538 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20167419 119 - 23771897 AAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAGAGUCUAUAGUCUUGGACAAUGCAAGUGCAUGCCAUGUAAAUAGCUAUACAGAUAGCUAUAGCCGCGCAUUAUAUCGUA ...........((((.(((((.(((.....(((((((((..........))))))))).((((....)))))))......((((((((....))))))))....))))).))))..... ( -30.40) >DroSec_CAF1 4489 117 - 1 AAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAGGUCUAU--UCUUGGACUAUGCAAGUGCAUGCCAUGUAAAUAGCUAUACUGAUAGCUAUAGCCGCGCAUUAUAUCGUA ...........((((.(((((.(((...((((((((((..........--))))))))))(((....))).)))......((((((((....))))))))....))))).))))..... ( -30.20) >DroSim_CAF1 6834 117 - 1 AAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAGGUCUAU--UCUUGGACUAUGCAAGUGCAUGCCAUGUAAAUAGCUAUACUGAUAGCUAUAGCCGCGCAUUAUAUCGUA ...........((((.(((((.(((...((((((((((..........--))))))))))(((....))).)))......((((((((....))))))))....))))).))))..... ( -30.20) >DroEre_CAF1 4514 117 - 1 AAGUCACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUACAGUCUAU--UCUUGGACUAUGCAAGUGCAUGCCAUGUAAAUAGCUAUACAAAUAGCUAUAGCUGCGCAUUAUAUCGUA ...........((((.(((((((((...((((((((((..........--))))))))))(((....))).)))......((((((((....))))))))...)))))).))))..... ( -31.00) >DroYak_CAF1 4477 117 - 1 AAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAAGUCUAU--UCUCGGACUAUGCAAGUGCAUGCCAUGUAAAUAGCUAUACAAAUAGCUAUAGCCGCGCAUUAUAUCAUA ...........((((.(((((.(((...(((((..(((..........--)))..)))))(((....))).)))......((((((((....))))))))....))))).))))..... ( -28.20) >consensus AAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAAGUCUAU__UCUUGGACUAUGCAAGUGCAUGCCAUGUAAAUAGCUAUACAGAUAGCUAUAGCCGCGCAUUAUAUCGUA ...........((((.(((((.(((....(((((((((............)))))))))((((....)))))))......((((((((....))))))))....))))).))))..... (-27.50 = -27.90 + 0.40)

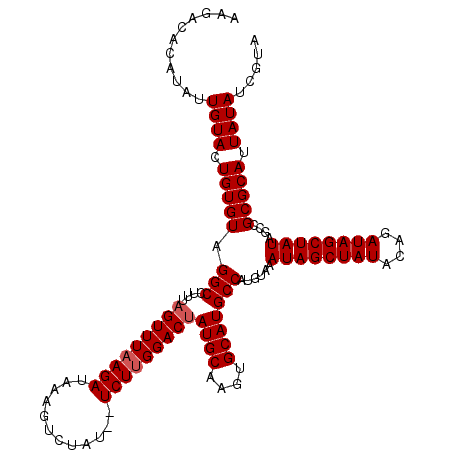
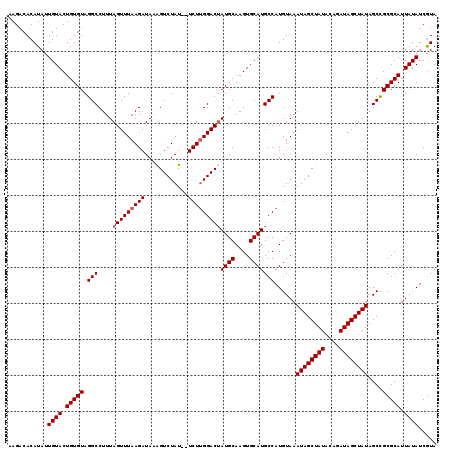
| Location | 20,167,459 – 20,167,571 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20167459 112 + 23771897 UACAUGGCAUGCACUUGCAUUGUCCAAGACUAUAGACUCUAUCUUAAACUAAAGGCCUACACAGUACAAUAUGUGUCUUGCACUAAAACUAAGCUGUUGCGUGUCUUCUGGU .....((((((((...((...(((..........)))...............((((..(((((........)))))...)).))........))...))))))))....... ( -21.20) >DroSec_CAF1 4529 110 + 1 UACAUGGCAUGCACUUGCAUAGUCCAAGA--AUAGACCUUAUCUUAAACUAAAGGCCUACACAGUACAAUAUGUGUCUUGCACUAAAACUAAGCUUUUGCGUGUCUUCUGGU .....((((((((((((..((((.(((((--.(((.((((...........)))).)))((((........))))))))).))))....))))....))))))))....... ( -25.50) >DroSim_CAF1 6874 110 + 1 UACAUGGCAUGCACUUGCAUAGUCCAAGA--AUAGACCUUAUCUUAAACUAAAGGCCUACACAGUACAAUAUGUGUCUUGCACUAAAACUAAGCUUUUGCGUGUCUUCUGGU .....((((((((((((..((((.(((((--.(((.((((...........)))).)))((((........))))))))).))))....))))....))))))))....... ( -25.50) >DroEre_CAF1 4554 110 + 1 UACAUGGCAUGCACUUGCAUAGUCCAAGA--AUAGACUGUAUCUUAAACUAAAGGCCUACACAGUACAAUAUGUGACUUGCACUAAAACUAAGCUUUUGCGUGUCUUCUGGU .....((((((((...((((((((.....--...))))))............((((...((((........))))....)).))........))...))))))))....... ( -24.00) >DroYak_CAF1 4517 110 + 1 UACAUGGCAUGCACUUGCAUAGUCCGAGA--AUAGACUUUAUCUUAAACUAAAGGCCUACACAGUACAAUAUGUGUCUUGCACUAAAACUAAGCUUUUGCGUGUCUUCUGGU .....((((((((...((.((((...((.--.....(((((........)))))((..(((((........)))))...)).))...)))).))...))))))))....... ( -26.20) >consensus UACAUGGCAUGCACUUGCAUAGUCCAAGA__AUAGACUUUAUCUUAAACUAAAGGCCUACACAGUACAAUAUGUGUCUUGCACUAAAACUAAGCUUUUGCGUGUCUUCUGGU .....((((((((...((.((((..((((............))))..)))).((((..(((((........)))))...)).))........))...))))))))....... (-22.20 = -22.44 + 0.24)



| Location | 20,167,459 – 20,167,571 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 20167459 112 - 23771897 ACCAGAAGACACGCAACAGCUUAGUUUUAGUGCAAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAGAGUCUAUAGUCUUGGACAAUGCAAGUGCAUGCCAUGUA ........((((((....))........(((((((........))))))))))).(((.....(((((((((..........))))))))).((((....)))))))..... ( -27.50) >DroSec_CAF1 4529 110 - 1 ACCAGAAGACACGCAAAAGCUUAGUUUUAGUGCAAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAGGUCUAU--UCUUGGACUAUGCAAGUGCAUGCCAUGUA ........(((.(((...((((.((..((((.(((((((((........))))((((((((...........)))))))).--))))).)))).))))))...)))..))). ( -28.80) >DroSim_CAF1 6874 110 - 1 ACCAGAAGACACGCAAAAGCUUAGUUUUAGUGCAAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAGGUCUAU--UCUUGGACUAUGCAAGUGCAUGCCAUGUA ........(((.(((...((((.((..((((.(((((((((........))))((((((((...........)))))))).--))))).)))).))))))...)))..))). ( -28.80) >DroEre_CAF1 4554 110 - 1 ACCAGAAGACACGCAAAAGCUUAGUUUUAGUGCAAGUCACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUACAGUCUAU--UCUUGGACUAUGCAAGUGCAUGCCAUGUA ........((((((....))........(((((((........))))))))))).(((...((((((((((..........--))))))))))(((....))).)))..... ( -27.30) >DroYak_CAF1 4517 110 - 1 ACCAGAAGACACGCAAAAGCUUAGUUUUAGUGCAAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAAGUCUAU--UCUCGGACUAUGCAAGUGCAUGCCAUGUA ........((((((....))........(((((((........))))))))))).(((...(((((..(((..........--)))..)))))(((....))).)))..... ( -25.40) >consensus ACCAGAAGACACGCAAAAGCUUAGUUUUAGUGCAAGACACAUAUUGUACUGUGUAGGCCUUUAGUUUAAGAUAAAGUCUAU__UCUUGGACUAUGCAAGUGCAUGCCAUGUA ........((((((....))........(((((((........))))))))))).(((....(((((((((............)))))))))((((....)))))))..... (-24.66 = -25.06 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:57 2006