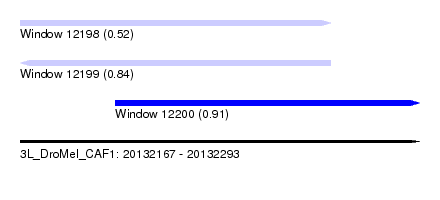
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 20,132,167 – 20,132,293 |
| Length | 126 |
| Max. P | 0.907511 |

| Location | 20,132,167 – 20,132,265 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -15.01 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
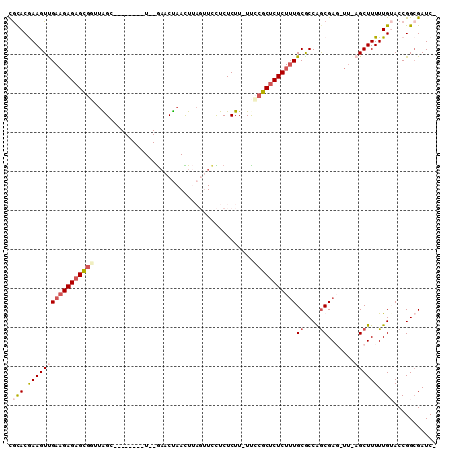
>3L_DroMel_CAF1 20132167 98 + 23771897 -GAUCGCCGGUACAAAAAGCUUAAUCUCGUUGGCGCAAAGAGAGCGGGA-AAGAGAGGAACUAAGUUAGUUC--A--------CCUAACCGCUCUCUUCAACUUCGUGCG -...((((((........(((..........)))...((((((((((..-...((..((((((...))))))--.--------.))..)))))))))).....))).))) ( -31.30) >DroSec_CAF1 46353 98 + 1 -GAUCGCCGGUACAAAAAGCUCAAGCUCGCCGGUGCAAAGAGAGCGGAA-AAGAGAGGAACUAAGUUAGUUC--A--------ACUAGUCGCUCUCUUCAACUUCGUGCG -....((((((......(((....))).))))))(((((((((((((..-...((..((((((...))))))--.--------.))..))))))))))........))). ( -32.70) >DroSim_CAF1 46530 98 + 1 -GAUCGCCGGUACAAAAAGCUCAAGCUCGCUGGCGCAAAGAGAGCGGGA-AAGAGAGGAACUAAGUUAGUUC--A--------ACUAACCGCUCUCUUCAACUUCGUGCG -....((((((......(((....))).))))))(((((((((((((..-...((..((((((...))))))--.--------.))..))))))))))........))). ( -35.70) >DroEre_CAF1 48370 95 + 1 UGAUCUCCGGUCCAAAAAGC------UCGCUGGUGCAAAGAGCGCGGAAGAAGAGAAAA----AAUUGGUGUUUAAGAGAAGAGCC----ACUCUC-UCGACUUUGUACA .......((((.........------..))))(((((((((((((.((...........----..)).)))))...((((.(((..----.)))))-))..)))))))). ( -20.22) >DroYak_CAF1 50746 99 + 1 UGAUCGCCGGUCCAAAAAGC------UCGCUGGCGCA-AGAGAGCGCAAAAAGAGAACA----AAUUAGAUCUAAGAAGAAGAGCCAUGCACUCUCUUCGACUUUGUGUG ....(((((((.........------..)))))))..-.....(((((((...(((.(.----.....).)))..(((((.(((.......))))))))...))))))). ( -23.50) >consensus _GAUCGCCGGUACAAAAAGCU_AA_CUCGCUGGCGCAAAGAGAGCGGAA_AAGAGAGGAACUAAGUUAGUUC__A________ACUAACCGCUCUCUUCAACUUCGUGCG ....(((((((.................)))))))..((((((((((.........................................))))))))))............ (-15.01 = -15.85 + 0.84)

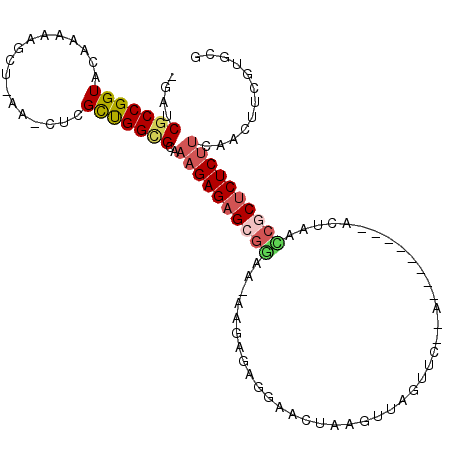
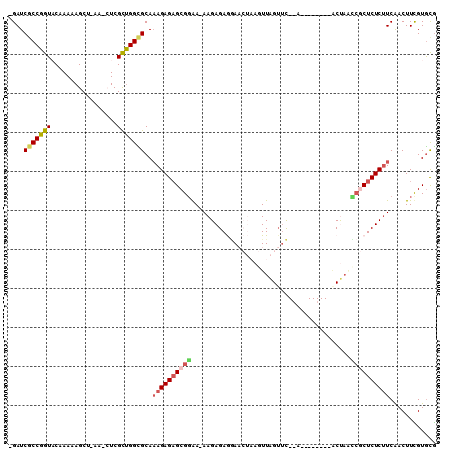
| Location | 20,132,167 – 20,132,265 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -14.50 |
| Energy contribution | -15.90 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
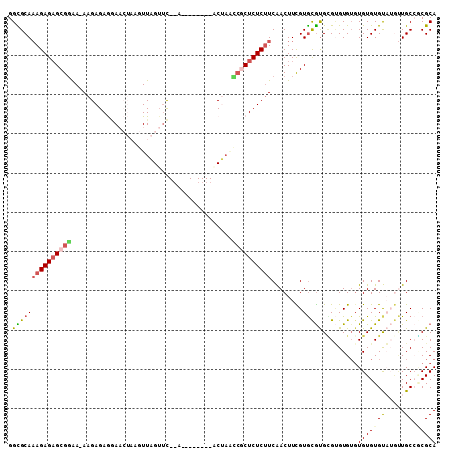
>3L_DroMel_CAF1 20132167 98 - 23771897 CGCACGAAGUUGAAGAGAGCGGUUAGG--------U--GAACUAACUUAGUUCCUCUCUU-UCCCGCUCUCUUUGCGCCAACGAGAUUAAGCUUUUUGUACCGGCGAUC- ...........(((((((((((..(((--------.--((((((...))))))....)))-..))))))))))).((((.((((((.......))))))...))))...- ( -34.60) >DroSec_CAF1 46353 98 - 1 CGCACGAAGUUGAAGAGAGCGACUAGU--------U--GAACUAACUUAGUUCCUCUCUU-UUCCGCUCUCUUUGCACCGGCGAGCUUGAGCUUUUUGUACCGGCGAUC- (((.((..(..((((((((.(((((((--------(--(...)))).))))).)))))))-)..)((((..(((((....)))))...)))).........)))))...- ( -32.40) >DroSim_CAF1 46530 98 - 1 CGCACGAAGUUGAAGAGAGCGGUUAGU--------U--GAACUAACUUAGUUCCUCUCUU-UCCCGCUCUCUUUGCGCCAGCGAGCUUGAGCUUUUUGUACCGGCGAUC- (((.((.....(((((((((((..((.--------.--((((((...))))))....)).-..)))))))))))....(((.((((....)))).)))...)))))...- ( -33.40) >DroEre_CAF1 48370 95 - 1 UGUACAAAGUCGA-GAGAGU----GGCUCUUCUCUUAAACACCAAUU----UUUUCUCUUCUUCCGCGCUCUUUGCACCAGCGA------GCUUUUUGGACCGGAGAUCA ...........((-(((((.----.....)))))))...........----...(((((...((((.((((...((....))))------))....))))..)))))... ( -19.00) >DroYak_CAF1 50746 99 - 1 CACACAAAGUCGAAGAGAGUGCAUGGCUCUUCUUCUUAGAUCUAAUU----UGUUCUCUUUUUGCGCUCUCU-UGCGCCAGCGA------GCUUUUUGGACCGGCGAUCA ........((((((((((((.....)))).)))))...(.(((((..----.((((.((....((((.....-.)))).)).))------))...))))).))))..... ( -27.90) >consensus CGCACGAAGUUGAAGAGAGCGGUUAGC________U__GAACUAACUUAGUUCCUCUCUU_UUCCGCUCUCUUUGCGCCAGCGAG_UU_AGCUUUUUGUACCGGCGAUC_ .(((.(((((((((((((((((.........................................)))))))))))((....)).......)))))).)))........... (-14.50 = -15.90 + 1.40)

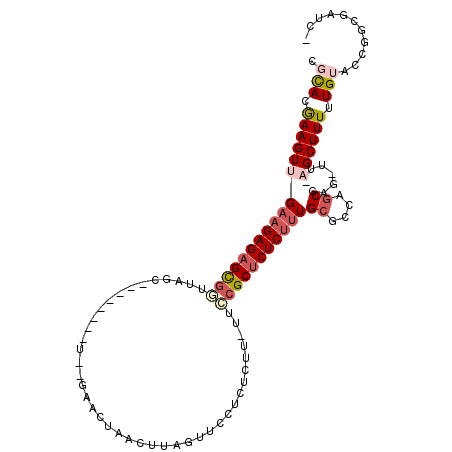
| Location | 20,132,197 – 20,132,293 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.32 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
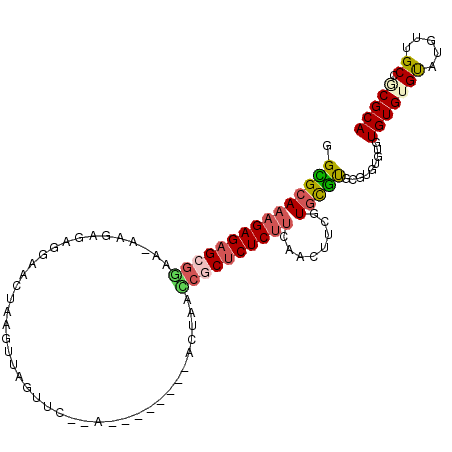
>3L_DroMel_CAF1 20132197 96 + 23771897 GGCGCAAAGAGAGCGGGA-AAGAGAGGAACUAAGUUAGUUC--A--------CCUAACCGCUCUCUUCAACUUCGUGCGUGCGUGUG--UGUGUGUAUGUUGCCGCGCA .((((.((((((((((..-...((..((((((...))))))--.--------.))..))))))))))......((((((..((....--))..)))))).....)))). ( -41.00) >DroSec_CAF1 46383 98 + 1 GGUGCAAAGAGAGCGGAA-AAGAGAGGAACUAAGUUAGUUC--A--------ACUAGUCGCUCUCUUCAACUUCGUGCGUGAGUGUGUGUGUGUGUAUGUUGCCUCGCA .(((((((((((((((..-...((..((((((...))))))--.--------.))..))))))))))..((..((..(....)..))...)).)))))........... ( -30.90) >DroSim_CAF1 46560 98 + 1 GGCGCAAAGAGAGCGGGA-AAGAGAGGAACUAAGUUAGUUC--A--------ACUAACCGCUCUCUUCAACUUCGUGCGUGCGUGUGUGUGUGUGUAUGUUGCCUCGCA .(((..((((((((((..-...((..((((((...))))))--.--------.))..))))))))))((((..((..((..(....)..))..))...))))...))). ( -37.10) >DroEre_CAF1 48395 96 + 1 GGUGCAAAGAGCGCGGAAGAAGAGAAAA----AAUUGGUGUUUAAGAGAAGAGCC----ACUCUC-UCGACUUUGUACAUGUGCGUGUGUGUGUGC----UGCGGCGCA .((((...(((((((((((..((((...----...((((.(((.....))).)))----)...))-))..)))).(((((....)))))..)))))----).).)))). ( -25.80) >DroYak_CAF1 50771 104 + 1 GGCGCA-AGAGAGCGCAAAAAGAGAACA----AAUUAGAUCUAAGAAGAAGAGCCAUGCACUCUCUUCGACUUUGUGUGCGUGAGUGUGUGUGUGCUGACUGCGACGCA .(((((-.....(((((.(((((((.(.----.....).)))..(((((.(((.......))))))))..)))).))))).....)))))((((((.....)).)))). ( -30.40) >consensus GGCGCAAAGAGAGCGGAA_AAGAGAGGAACUAAGUUAGUUC__A________ACUAACCGCUCUCUUCAACUUCGUGCGUGCGUGUGUGUGUGUGUAUGUUGCCGCGCA .(((((((((((((((.........................................))))))))))........))))).........(((((((.....)).))))) (-14.04 = -14.68 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:52 2006